Systematic screening of metabolic pathways to identify two breast cancer subtypes with divergent immune characteristics
- PMID: 40594370
- PMCID: PMC12216186
- DOI: 10.1038/s41598-025-05179-7
Systematic screening of metabolic pathways to identify two breast cancer subtypes with divergent immune characteristics
Abstract
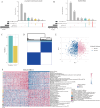
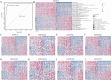
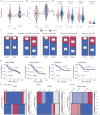
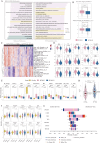
Due to the high heterogeneity among breast cancer (BRCA) patients, most individuals show a limited response rate to one specific treatment. The metabolic plasticity of BRCA cells is one of the main causes of their heterogeneity, affecting not only their own growth and function but also their metabolites have an impact on the tumor immune microenvironment (TIME). However systematic evaluation of metabolic pathways in BRCA is lacking. We identified BRCA metabolic subtypes (BCMS) by consensus clustering 26 KEGG/Reactome pathways in the TCGA BRCA discovery cohort (n = 1094). Nine independent bulk transcriptome cohorts (total n > 4000), including METABRIC and GEO datasets, were used for validation via random forest classification. To characterize BCMS, we applied an analytical framework encompassing functional enrichment (GSEA), immune infiltration (Mcpcounter), clinical correlation, drug sensitivity (oncoPredict) on bulk transcriptome data, cell-cell communication analysis (CellChat) on single-cell RNA sequencing (scRNA-seq) data, and spatial co-localization analysis (CellTrek) on spatial RNA sequencing (spRNA-seq) data. We identified two distinct BCMS. BCMS-I exhibited upregulated lipid metabolism-related pathways, characterized by immune activation, a better prognosis, and higher infiltration of immune cells, including B cells, T cells, NK cells, macrophages, and neutrophils. Spatial co-localization analysis further revealed that BCMS-I demonstrated spatial co-localization with immune cells. In contrast, BCMS-II showed upregulation of amino acid and vitamin metabolism-related pathways, with tumor cell proliferation, a poorer prognosis, and a lack of immune cell infiltration. The immune activation in BCMS-I is marked by the significant activation of the MHC-I signaling pathway in interactions between tumor cells and T/NK cells, and of the MHC-II signaling pathway in interactions between tumor cells and dendritic cells/macrophages. In contrast, the proliferative characteristics of BCMS-II are associated with the co-activation of the GRN signaling pathway by myeloid immune cells and stromal cells within the tumor microenvironment. Drug sensitivity analysis revealed that BCMS-II was highly sensitive to Ganitumab, Carboplatin + ABT-888, and Pembrolizumab. This study established a novel Breast Cancer Metabolic Subtyping System (BCMSS) based on metabolic pathway analysis. Our findings highlight the heterogeneity of BRCA in terms of metabolic features, immune characteristics, clinical prognosis, and drug sensitivity. The novel classification system provides valuable insights for clinical diagnosis and treatment, serving as a foundation for precision diagnosis and personalized therapies in BRCA.
Keywords: Breast cancer; Metabolic plasticity; Metabolic subtypes; Tumor immune microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Integrated proteomics and transcriptomics analysis reveals key regulatory genes between ER-positive/PR-positive and ER-positive/PR-negative breast cancer.BMC Cancer. 2025 Jul 1;25(1):1048. doi: 10.1186/s12885-025-14451-y. BMC Cancer. 2025. PMID: 40597935 Free PMC article.
-
Deciphering the tumor immune microenvironment: single-cell and spatial transcriptomic insights into cervical cancer fibroblasts.J Exp Clin Cancer Res. 2025 Jul 5;44(1):194. doi: 10.1186/s13046-025-03432-5. J Exp Clin Cancer Res. 2025. PMID: 40616092 Free PMC article.
-
Integrated single-cell and transcriptomic analysis of bone marrow-derived metastatic neuroblastoma reveals molecular mechanisms of metabolic reprogramming.Sci Rep. 2025 Aug 5;15(1):28519. doi: 10.1038/s41598-025-13626-8. Sci Rep. 2025. PMID: 40764361 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

