Temperature abuse and Salmonella Typhimurium colonization disrupt the indigenous bacterial communities of pasteurized bovine milk over time
- PMID: 40594414
- PMCID: PMC12219400
- DOI: 10.1038/s41598-025-06838-5
Temperature abuse and Salmonella Typhimurium colonization disrupt the indigenous bacterial communities of pasteurized bovine milk over time
Abstract
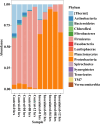
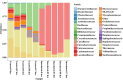
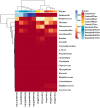
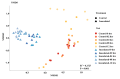
Even though the population structure of the bovine milk residential bacterial population is known, the alterations in the population structure associated with food safety issues, such as temperature abuse/pathogen colonization, are unknown. Here, alterations of the bacterial population, either incubated at 37 °C (temperature abuse) or inoculated with Salmonella Typhimurium (pathogen colonization), were characterized using full-length 16S rRNA sequencing. At zero hour, the bacterial population of milk primarily constituted of Firmicutes and Thermi. Of the 218 genera identified, Thermus (37%) and Streptococcus (34%) were the most dominant. The 12-h incubation at 37 °C replaced almost 96% of the population by Firmicutes, exemplified by a remarkable increase in the abundance of the genus Bacillus. Concurrently, only 36 genera survived, with an abundance of Bacillus, which showed a 98-fold increase during the 12-h incubation. Similarly, only 150 genera remained after 12 h in Salmonella-inoculated milk. Both temperature abuse and Salmonella inoculation significantly reduce bacterial diversity and richness. Nonmetric multidimensional scaling analysis between the control and Salmonella inoculated samples was significantly distinct at all times, confirming alterations in the bacterial population during Salmonella colonization. Even though the load of Firmicutes increased temporally, bacteria belonging to the genera Bacillus, Macrococcus, and Serratia genera were depleted in Salmonella inoculated milk samples. Taken together, both experimental conditions, viz. temperature abuse and Salmonella contamination, demonstrated a significant drop in residential milk bacterial diversity. This general drop in bacterial diversity could allow Salmonella to occupy and colonize the milk matrix.
Keywords: Salmonella Typhimurium; 16S rRNA amplicon sequencing; Bacterial community; Bovine milk; Milk microbiology.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Viable bacterial communities in freshly pumped human milk and their changes during cold storage conditions.Int Breastfeed J. 2025 May 29;20(1):44. doi: 10.1186/s13006-025-00738-0. Int Breastfeed J. 2025. PMID: 40442718 Free PMC article.
-
Characteristics of the milk microbiota of healthy goats and goats diagnosed with clinical mastitis in Western China.Microb Pathog. 2025 Sep;206:107764. doi: 10.1016/j.micpath.2025.107764. Epub 2025 Jun 1. Microb Pathog. 2025. PMID: 40460985
-
Characterization of bacterial and fungal populations in retail kefirs in Ireland.J Dairy Sci. 2025 Aug;108(8):8187-8204. doi: 10.3168/jds.2025-26587. Epub 2025 May 28. J Dairy Sci. 2025. PMID: 40447085
-
Systematic Review of the Human Milk Microbiota.Nutr Clin Pract. 2017 Jun;32(3):354-364. doi: 10.1177/0884533616670150. Epub 2016 Sep 27. Nutr Clin Pract. 2017. PMID: 27679525
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Quigley, L. et al. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J. Dairy Sci.96, 4928–4937 (2013). - PubMed
-
- Ding, R. et al. Characterization of the core microflora and nutrient composition in packaged pasteurized milk products during storage. Food Sci. Human Wellness12, 1279–1286 (2023).
-
- Derakhshani, H., Plaizier, J. C., De Buck, J., Barkema, H. W. & Khafipour, E. Composition of the teat canal and intramammary microbiota of dairy cows subjected to antimicrobial dry cow therapy and internal teat sealant. J. Dairy Sci.101, 10191–10205 (2018). - PubMed
-
- Masoud, W. et al. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. Int. Dairy J.21, 142–148 (2011).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

