Development of a candidate mRNA vaccine based on Multi-Peptide targeting VP4 of rotavirus A: an immunoinformatics and molecular dynamics approach
- PMID: 40594603
- PMCID: PMC12218802
- DOI: 10.1038/s41598-025-07433-4
Development of a candidate mRNA vaccine based on Multi-Peptide targeting VP4 of rotavirus A: an immunoinformatics and molecular dynamics approach
Abstract
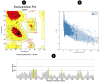
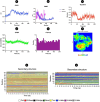
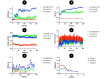
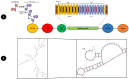
Rotavirus (RV) is a common double-stranded RNA virus that causes diarrheal disease in young children. The prevalent species, Rotavirus A (RVA), is responsible for over 90% of human RV infections. With significant morbidity and mortality, this pathogen poses a serious global health challenge, particularly in underdeveloped countries. This study presents an immunoinformatics approach for designing an mRNA vaccine based on a multi-peptide construct to elicit robust immune responses against RVA. The VP4 was analyzed from 40 sequences using phylogenetic analysis. Prediction of cytotoxic (CTL) and helper T cell (HTL) epitopes was performed and validated. The 17 high-conservancy CTL/HTL epitopes were selected for vaccine construction. The mRNA vaccine based on multi-peptide was engineered with human beta-defensin 3 (hBD3) adjuvant and linkers to enhance immunogenicity. The designed mRNA vaccine product exhibited favorable physicochemical properties and was predicted to be a probable antigen, non-allergenic, and non-toxic. 2D and 3D structure validation demonstrated the quality of the model. Molecular docking with Toll-like receptor 2/3 (TLR2/3) indicated favorable interaction, and peptide docking with MHC-I/II alleles showed strong binding affinities and have significant Residue-Residue interactions. Simulation of immune responses revealed potent B-cell and T-cell activities, macrophage responses, and significant cytokine synthesis. Molecular dynamics simulation (MDS) confirmed the structural stability of the TLR3-vaccine complex, and MHC-peptide in 200ns and STQFTDFVSLNSLRF peptide have shown good interaction with MHC molecule. In addition, the MM/GBSA analysis yielded a binding free energy of - 89.77 kcal/mol, indicating a strong and stable interaction between the vaccine construct and the target receptor. Codon optimization and mRNA secondary structure prediction were carried out for efficient translation. Additionally, population coverage analysis indicated the vaccine's effectiveness worldwide with 100% value. Overall, this study showcases a promising immunoinformatics approach for designing an mRNA vaccine based on a multi-peptide construct targeting RVA. The findings support the potential of this vaccine design to elicit robust and widespread immune responses against RVA infection, paving the way for future vaccine development strategies and this study needs experimental validation.
Keywords: Computational immunology; Immunoinformatics; Molecular dynamics simulation; Multi-peptide; Rotavirus A; mRNA vaccine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
-
- Lekana-Douki, S. E. et al. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. Int. J. Infect. Dis. IJID Off Publ Int. Soc. Infect. Dis.34, 90–95 (2015). - PubMed
-
- on Taxonomy of Viruses, I. C. & & King, A. M. Q. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses (Elsevier Science, 2011).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

