A bitopic mTORC inhibitor reverses phenotypes in a tuberous sclerosis complex model
- PMID: 40594897
- PMCID: PMC12218406
- DOI: 10.1038/s41598-025-08345-z
A bitopic mTORC inhibitor reverses phenotypes in a tuberous sclerosis complex model
Abstract
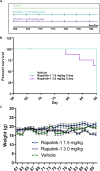
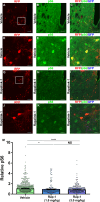
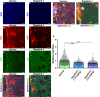
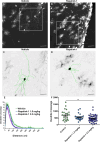
Neural stem cells (NSCs) of the ventricular-subventricular zone (V-SVZ) generate diverse cell types including striatal glia during the neonatal period. NSC progeny uncouple stem cell-related mRNA transcripts from being translated during differentiation. We previously demonstrated that Tsc2 inactivation, which occurs in the neurodevelopmental disorder Tuberous Sclerosis Complex (TSC), prevents this from happening. Loss of Tsc2 causes hyperactivation of the protein kinase mechanistic target of rapamycin complex 1 (mTORC1), altered translation, retention of stemness in striatal glia, and the production of misplaced cytomegalic neurons having hypertrophic dendrite arbors. These phenotypes model characteristics of TSC hamartomas called subependymal giant cell astrocytomas (SEGAs). mTORC1 inhibitors called rapamycin analogs (rapalogs) are currently used to treat TSC and to assess the role of mTORC1 in regulating TSC-related phenotypes. Rapalogs are useful for treating SEGAs. However, they require lifelong application, have untoward side effects, and resistance may occur. They also incompletely inhibit mTORC1 and have limited efficacy. Rapalink-1 is a bitopic inhibitor that links rapamycin to a second-generation mTOR ATP competitive inhibitor, MLN0128. Here we explored the effect of Rapalink-1 on a TSC hamartoma model. The model is created by neonatal electroporation of mice having conditional Tsc2 genes. Prolonged Rapalink-1 treatment could be achieved with 1.5 or 3.0 mg/Kg injected intraperitoneally every five days. Rapalink-1 inhibited the mTORC1 pathway, decreased cell size, reduced neuron dendrite arbors, and reduced hamartoma size. In conclusion, these results demonstrate that cellular phenotypes in a TSC SEGA model are reversed by Rapalink-1 which may be useful to resolve TSC brain hamartomas.
Keywords: MTORC1; Neurogenesis; Rapalink-1; SEGA; Subependymal giant cell Astrocytoma; Subependymal nodule; TSC; Tsc2; Tuberous sclerosis complex.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Conflict of interest: The authors declare that the researchers have no competing financial interests.
Figures

References
-
- Northrup, H. et al. Updated international tuberous sclerosis complex diagnostic criteria and surveillance and management recommendations. Pediatr. Neurol.12310.1016/j.pediatrneurol.2021.07.011 (2021). - PubMed
-
- Feliciano, D. M. Modeling genetic mosaicism of the mammalian target of rapamycin pathway in the cerebral cortex. Front. Mammal Sci. 2. (2023). 10.3389/fmamm.2023.1231778
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

