Site- and cell-type-specific miRNA and mRNA genes and networks across the cortex, striatum, and hypothalamus
- PMID: 40594985
- PMCID: PMC12218356
- DOI: 10.1038/s42003-025-08371-7
Site- and cell-type-specific miRNA and mRNA genes and networks across the cortex, striatum, and hypothalamus
Abstract
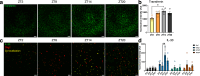
Biological rhythms control gene expression, but effects on central nervous system (CNS) cells and structures remain poorly defined. While circadian (24-hour) rhythms are most studied, many genes have periods of greater and less than 24-hours; these fluctuations can be both site- and cell-specific. Identifying patterns of gene rhythmicity across the CNS is necessary for both the study of chronobiology and to make sense of data obtained in the laboratory. We now identify cycling mRNAs, miRNAs, gene networks and mRNA-miRNA co-expression pairs in the cortex, hypothalamus, and corpus striatum of male C57BL/6J mice using high-dimensional datasets. A searchable catalogue ( https://www.ghasemloulab.ca/chronoCNS ) helps refine the analysis of cellular and molecular rhythmicity across the CNS (using the liver as a control). Immunofluorescence confirms the rhythmicity of key targets across cells in these structures, with strong cycling signatures in resting oligodendrocytes. Our study sheds light on the contribution of diurnal, ultradian, and infradian rhythms and mRNA-miRNA interactions to CNS function.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.G.T. is currently an employee of Olink Proteomics AB, however, the published work was done prior to this employment and does not involve/promote any of Olink’s materials or point of view. H.C. is currently an employee of Geneseeq Technology Inc., however, the published work was done prior to this employment and does not involve/promote any of Geneseeq’s materials or points of view. H.G. is currently an employee at Caruta Therapeutics, however, the published work was done prior to this employment and does not involve/promote any of Caruta’s materials or points of view. All other authors declare no competing interests.
Figures









References
MeSH terms
Substances
Grants and funding
- RGPIN-05604/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- PJT-190170/Gouvernement du Canada | Instituts de Recherche en Santé du Canada | CIHR Skin Research Training Centre (Skin Research Training Centre)
LinkOut - more resources
Full Text Sources

