Metagenomic analysis to identify unique microbes in the rhizosphere of basmati rice (Oryza sativa L.) accessions
- PMID: 40595004
- PMCID: PMC12218860
- DOI: 10.1038/s41598-025-87889-6
Metagenomic analysis to identify unique microbes in the rhizosphere of basmati rice (Oryza sativa L.) accessions
Abstract
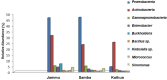
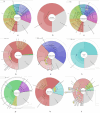
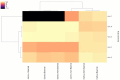
The captivating aroma of basmati rice is highly favoured by consumers across the globe. Unfortunately, the aroma of basmati rice has been gradually diminishing over time due to the excessive use of inorganic fertilizers and the impact of climate change. To understand the microbial community that plays a significant role in aroma enhancement in basmati rice accessions, a systematic study is required. A unique rhizobacteria of basmati rice associated with basmati rice were Actinobacteria, Bacillus subtilis, Burkholderia, Enterobacter, Klebsiella, Lactobacillus, Micrococcus, Pseudomonas, and Sinomonas. The biosynthesis of potential precursors (ornithine, putrescine, proline, and polyamines) of aroma in basmati rice involved various enzymes such as acetylornithine aminotransferase, acetylornithine deacetylase, N-acetylornithine carbomyltransferase, acetylornithine/succinyldiaminopimelate aminotransferase, and ornithine cyclodeaminase. These findings significantly contribute to the existing understanding of the rhizobacteria associated with basmati rice that play a crucial role in enhancing the aroma. The introduction of these cultures into the basmati rice growing areas has the potential to augment the plant growth and enhances the aroma. The present study explored the functional potential of the microbial community associated with aroma improvement in basmati rice. This will also enhance the export potential of the basmati rice in the region on sustainable basis.
Keywords: Aroma enhancement; Basmati rice; Geographical indications (GIs); Metagenomics; Rhizobacteria.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: The collection of rice rhizosphere resources and research activities has been conducted in compliance with the Regulations on Resident Instructions and duly approved by the Competent Authority of Sher-e-Kashmir University of Agricultural Sciences and Technology of Jammu, Main Campus, Chatha, Jammu, India.
Figures
References
-
- Okpala, N. E., Mo, Z., Duan, M. & Tang, X. The genetics and biosynthesis of 2-acetyl-1-pyrroline in fragrant rice. Plant Physiol. Biochem.135, 272–276. 10.1016/j.plaphy.2018.12.012 (2019). - PubMed
-
- Yoshihashi, T., Huong, N. T. & Inatomi, H. Precursors of 2-acetyl-1-pyrroline, a potent flavor compound of an aromatic rice variety. J. Agric. Food Chem.50(7), 2001–2004. 10.1021/jf011268s.PMID:11902947 (2002). - PubMed
-
- Salgotra, R.K., Bhat, J.A., Gupta, B.B. & Sharma, S. Determination of genetic relationship among basmati and non-basmati rice (Oryza sativa L.) genotypes from North-West Himalayas using microsatellite markers. Indian J. Biotech. 16, 68–75 (2017).
-
- Singh, R. et al. Aromatic Rices. Oxford and IBH; New Delhi, India. Small and Medium Grained Aromatic Rices of India; pp. 155–177 (2000).
MeSH terms
LinkOut - more resources
Full Text Sources

