Comparison of sepsis-associated acute kidney injury with different degrees and causes reveals patterns in mitochondrial metabolism and immune infiltration changes
- PMID: 40595246
- PMCID: PMC12216226
- DOI: 10.1038/s41598-025-08192-y
Comparison of sepsis-associated acute kidney injury with different degrees and causes reveals patterns in mitochondrial metabolism and immune infiltration changes
Abstract
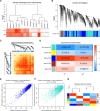
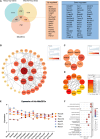
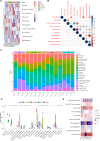
Sepsis-associated acute kidney injury (SA-AKI) is a common complication in critically ill patients, characterized by high morbidity and mortality rates. It remains primarily treated with supportive and nonspecific therapies because of the absence of effective diagnostic biomarkers and therapeutic targets. This study utilized cecal ligation and puncture (CLP) and intraperitoneal injection of lipopolysaccharide (LPS) to create a murine model of SA-AKI, simulating various degrees of severity and aetiologies. Renal transcriptome sequencing was performed, followed by extensive bioinformatic analyses, including gene expression trend analysis using the Mfuzz package, weighted gene co-expression network analysis (WGCNA), protein-protein interaction (PPI) networks, and Cibersort immune infiltration analysis, to uncover key molecular changes in SA-AKI. Transcriptome analysis revealed that SA-AKI induces mitochondrial dysfunction and metabolic disturbances. As the severity of SA-AKI increases, immune-inflammatory activity becomes more pronounced, along with significant metabolic dysfunction. Variations in immune cell infiltration between different aetiologies of SA-AKI suggest distinct immune response patterns and timing. Six hub mitochondrial differentially expressed genes (MitoDEGs) related to SA-AKI severity were identified and validated, showing significant associations with immune cell infiltration. These findings provide valuable insights into SA-AKI pathogenesis and the exploration of therapeutic targets.
Keywords: Bioinformatics analysis; Immune infiltration; Metabolism; Mitochondria; Molecular mechanism; Sepsis-associated acute kidney injury.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interest: The authors declare no competing interests. Ethical approval: The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Chinese PLA General Hospital, and the permit number is 2019-X15-97. This study was also conducted in accordance with the guidelines of the Animal Research: Reporting of In Vivo Experiments (ARRIVE).
Figures






Similar articles
-
Sepsis induces heterogeneous transcription of coagulation- and inflammation-associated genes in renal microvasculature.Thromb Res. 2024 May;237:112-128. doi: 10.1016/j.thromres.2024.03.014. Epub 2024 Mar 18. Thromb Res. 2024. PMID: 38579513
-
PLK3-Activated Mitochondrial Apoptosis Pathway in the Pathogenesis of Sepsis-Associated Acute Kidney Injury.J Biochem Mol Toxicol. 2025 Sep;39(9):e70462. doi: 10.1002/jbt.70462. J Biochem Mol Toxicol. 2025. PMID: 40874686
-
Mitochondrial RNA/RIG-I promotes Caspase-1/GSDMD-mediated inflammation in sepsis-associated acute kidney injury.Immunobiology. 2025 Jul;230(4):153085. doi: 10.1016/j.imbio.2025.153085. Epub 2025 Jun 4. Immunobiology. 2025. PMID: 40505524
-
Acute kidney injury following multisystem inflammatory syndrome associated with SARS-CoV-2 infection in children: a systematic review and meta-analysis.Pediatr Nephrol. 2023 Feb;38(2):357-370. doi: 10.1007/s00467-022-05701-3. Epub 2022 Aug 9. Pediatr Nephrol. 2023. PMID: 35943577 Free PMC article.
-
Intensity of continuous renal replacement therapy for acute kidney injury.Cochrane Database Syst Rev. 2016 Oct 4;10(10):CD010613. doi: 10.1002/14651858.CD010613.pub2. Cochrane Database Syst Rev. 2016. PMID: 27699760 Free PMC article.
References
-
- Zarbock, A. et al. Sepsis-associated acute kidney injury: Consensus report of the 28th acute disease quality initiative workgroup. Nat. Rev. Nephrol.19(6), 401–417. 10.1038/s41581-023-00683-3 (2023). - PubMed
-
- Hoste, E. A. J. et al. Epidemiology of acute kidney injury in critically ill patients: The multinational AKI-EPI study. Intensive Care Med.41(8), 1411–1423. 10.1007/s00134-015-3934-7 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

