Human gut bacteria bioaccumulate per- and polyfluoroalkyl substances
- PMID: 40595288
- PMCID: PMC12222025
- DOI: 10.1038/s41564-025-02032-5
Human gut bacteria bioaccumulate per- and polyfluoroalkyl substances
Abstract
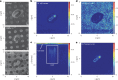
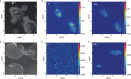
Per- and polyfluoroalkyl substances (PFAS) are persistent pollutants that pose major environmental and health concerns. While few environmental bacteria have been reported to bind PFAS, the interaction of PFAS with human-associated gut bacteria is unclear. Here we report the bioaccumulation of PFAS by 38 gut bacterial strains ranging in concentration from nanomolar to 500 μM. Bacteroides uniformis showed notable PFAS accumulation resulting in millimolar intracellular concentrations while retaining growth. In Escherichia coli, bioaccumulation increased in the absence of the TolC efflux pump, indicating active transmembrane transport. Cryogenic focused ion beam secondary-ion mass spectrometry confirmed intracellular localization of the PFAS perfluorononanoic acid (PFNA) in E. coli. Proteomic and metabolomic analysis of PFNA-treated cells, and the mutations identified following laboratory evolution, support these findings. Finally, mice colonized with human gut bacteria showed higher PFNA levels in excreted faeces than germ-free controls or those colonized with low-bioaccumulating bacteria. Together, our findings uncover the high PFAS bioaccumulation capacity of gut bacteria.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.E.L. and K.R.P. are inventors in a patent application related to the findings presented in the paper (UK patent application number 2215307.6). A.E.L., J.E.D.T. and K.R.P. are co-founders of Cambiotics ApS. The other authors declare no conflicts of interest.
Figures













References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

