High-resolution detection of copy number alterations in single cells with HiScanner
- PMID: 40595464
- PMCID: PMC12214996
- DOI: 10.1038/s41467-025-60446-5
High-resolution detection of copy number alterations in single cells with HiScanner
Abstract
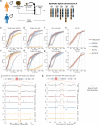
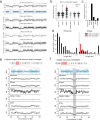
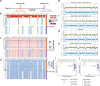
Improvements in single-cell whole-genome sequencing (scWGS) assays have enabled detailed characterization of somatic copy number alterations (CNAs) at the single-cell level. Yet, current computational methods are mostly designed for detecting chromosome-scale changes in cancer samples with low sequencing coverage. Here, we introduce HiScanner (High-resolution Single-Cell Allelic copy Number callER), which combines read depth, B-allele frequency, and haplotype phasing to identify CNAs with high resolution. In simulated data, HiScanner consistently outperforms state-of-the-art methods across various CNA types and sizes. When applied to high-coverage scWGS data from 65 cells across 11 neurotypical human brains, HiScanner shows a superior ability to detect smaller CNAs, uncovering distinct CNA patterns between neurons and oligodendrocytes. We also generated low-coverage scWGS data from 179 cells sampled from the same meningioma patient at two time points. For this serial dataset, integration of CNAs with point mutations revealed evolutionary trajectories of tumor cells. These findings show that HiScanner enables accurate characterization of frequency, clonality, and distribution of CNAs at the single-cell level in both non-neoplastic and neoplastic cells.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.J.P. is a member of the scientific advisory board (SAB) for Bioskryb Genomics, Inc. C.A.W. is a member of the SAB of Bioskryb Genomics, Inc. (cash, equity), Mosaica Therapeutics (cash, equity), and an advisor to Maze Therapeutics (equity). The remaining authors declare no competing interests.
Figures


Update of
-
High-resolution detection of copy number alterations in single cells with HiScanner.bioRxiv [Preprint]. 2025 Apr 3:2024.04.26.587806. doi: 10.1101/2024.04.26.587806. bioRxiv. 2025. Update in: Nat Commun. 2025 Jul 1;16(1):5477. doi: 10.1038/s41467-025-60446-5. PMID: 38746445 Free PMC article. Updated. Preprint.
References
-
- Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

