A constricted mitochondrial morphology formed during respiration
- PMID: 40595511
- PMCID: PMC12215465
- DOI: 10.1038/s41467-025-60658-9
A constricted mitochondrial morphology formed during respiration
Abstract
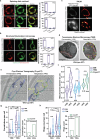
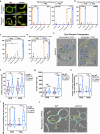
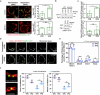
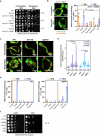
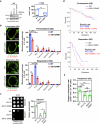
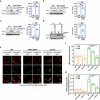
Mitochondria assemble in a dynamic tubular network. Their morphology is governed by mitochondrial fusion and fission, which regulate most mitochondrial functions including oxidative phosphorylation. Yet, the link between mitochondrial morphology and respiratgion remains unclear. Here, we uncover a mitochondrial morphology dedicated to respiratory growth of Saccharomyces cerevisiae, which we refer to as "Ringo". The Ringo morphology is characterized by stable constrictions of mitochondrial tubules. Ringo constrictions are mediated by the yeast dynamin Dnm1 and, unlike mitochondrial fission, occur in the absence of contacts with the endoplasmic reticulum. Our data show that blocking formation of the Ringo morphology correlates with decreased respiration, decreased expression of OXPHOS subunits and perturbed mitochondrial DNA distribution. These results open important perspectives about the link between mitochondrial form and function.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Mechanistic study of modulating mitochondrial fission and fusion to ameliorate neuropathic pain in mice.Sci Rep. 2025 May 4;15(1):15571. doi: 10.1038/s41598-025-99300-5. Sci Rep. 2025. PMID: 40320455 Free PMC article.
-
MyD88 polymerization and association to cellular membranes in a yeast heterologous model.Cell Mol Life Sci. 2025 Jul 25;82(1):288. doi: 10.1007/s00018-025-05827-1. Cell Mol Life Sci. 2025. PMID: 40711583 Free PMC article.
-
Completion of mitochondrial division requires the intermembrane space protein Mdi1/Atg44.J Cell Biol. 2023 Oct 2;222(10):e202303147. doi: 10.1083/jcb.202303147. Epub 2023 Aug 4. J Cell Biol. 2023. PMID: 37540145 Free PMC article.
-
Pharmacological treatments in panic disorder in adults: a network meta-analysis.Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Quintana-Cabrera, R. & Scorrano, L. Determinants and outcomes of mitochondrial dynamics. Mol. Cell83, 857–876 (2023). - PubMed
-
- Ozeir, M. & Cohen, M. M. From dynamin related proteins structures and oligomers to membrane fusion mediated by mitofusins. Biochim Biophys. Acta Bioenerg.1863, 148913 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- ANR-11-LABX-0011-DYNAMO/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-CE13-0026-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-19-CE11-0018/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-CE13-0026-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-21-CE45-003-02/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources

