A large C-terminal Rad52 segment acts as a chaperone to Form and Stabilize Rad51 Filaments
- PMID: 40595518
- PMCID: PMC12218292
- DOI: 10.1038/s41467-025-60664-x
A large C-terminal Rad52 segment acts as a chaperone to Form and Stabilize Rad51 Filaments
Abstract
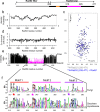
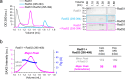
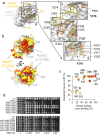
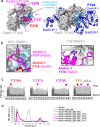
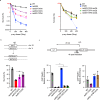
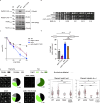
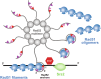
Homologous recombination (HR) is essential for the repair of DNA double-strand breaks and the restart of stalled replication forks. A critical step in HR is the formation of Rad51 nucleofilaments, which perform homology search and strand invasion of a homologous DNA sequence required for repair synthesis. In the yeast Saccharomyces cerevisiae, Rad52 facilitates Rad51 nucleofilament formation by mediating Rad51 loading onto ssDNA and counteracting Rad51 filament dissociation by the DNA translocase Srs2. The molecular basis of these two Rad52 functions remains unclear. Our integrative structural analyses of the Rad51-Rad52 interaction, combining NMR, SAXS, and modeling, reveal that an 85-residue segment of Rad52, conserved in fungi, folds upon binding to a broad surface of a Rad51 monomer. Notably, it includes an FxxA motif conserved in the BRC repeats of BRCA2 and at the Rad51-Rad51 interface. This binding mode was validated through an extensive set of mutations. Using in vivo assays and a functional fluorescent GFP-Rad51 fusion protein, we demonstrated that this entire segment is critical for Rad51 filament formation. These findings highlight how Rad52 functions as an assembly chaperone by preventing Rad51 oligomerization, promoting nucleation of Rad51 nucleofilaments on ssDNA, and counteracting the effects of Srs2 on destabilizing Rad51 filaments.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
- 2016-1-PL BIO-03-CEA-1/Institut National Du Cancer (French National Cancer Institute)
- ANR-20-CE18-0038/Agence Nationale de la Recherche (French National Research Agency)
- ANR-21-CE11-0027/Agence Nationale de la Recherche (French National Research Agency)
- ANR-23-CE11-0023/Agence Nationale de la Recherche (French National Research Agency)
- PGA1*20160203953/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

