Model-free photon analysis of diffusion-based single-molecule FRET experiments
- PMID: 40595536
- PMCID: PMC12216303
- DOI: 10.1038/s41467-025-60764-8
Model-free photon analysis of diffusion-based single-molecule FRET experiments
Abstract
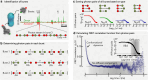
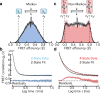
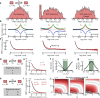
Photon-by-photon analysis tools for diffusion-based single-molecule Förster resonance energy transfer (smFRET) experiments often describe protein dynamics with Markov models. However, FRET efficiencies are only projections of the conformational space such that the measured dynamics can appear non-Markovian. Model-free methods to quantify FRET efficiency fluctuations would be desirable in this case. Here, we present such an approach. We determine FRET efficiency correlation functions free of artifacts from the finite length of photon trajectories or the diffusion of molecules through the confocal volume. We show that these functions capture the dynamics of proteins from nano- to milliseconds both in simulation and experiment, which provides a rigorous validation of current model-based analysis approaches.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Unraveling Burst Selection Bias in Single-Molecule FRET of Species with Unequal Brightness and Diffusivity.J Phys Chem B. 2024 Jun 13;128(23):5576-5589. doi: 10.1021/acs.jpcb.4c01178. Epub 2024 Jun 4. J Phys Chem B. 2024. PMID: 38833567
-
Three-color single-molecule maximum likelihood analysis of folding and binding of diffusing molecules.Biophys J. 2025 Sep 2;124(17):2909-2926. doi: 10.1016/j.bpj.2025.07.033. Epub 2025 Jul 30. Biophys J. 2025. PMID: 40745921 Free PMC article.
-
Quantitative analysis methods for free diffusion single-molecule FRET experiments.Curr Opin Struct Biol. 2025 Aug;93:103075. doi: 10.1016/j.sbi.2025.103075. Epub 2025 Jun 9. Curr Opin Struct Biol. 2025. PMID: 40494165 Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

