Model-free photon analysis of diffusion-based single-molecule FRET experiments
- PMID: 40595536
- PMCID: PMC12216303
- DOI: 10.1038/s41467-025-60764-8
Model-free photon analysis of diffusion-based single-molecule FRET experiments
Abstract
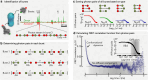
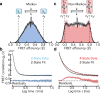
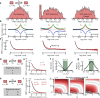
Photon-by-photon analysis tools for diffusion-based single-molecule Förster resonance energy transfer (smFRET) experiments often describe protein dynamics with Markov models. However, FRET efficiencies are only projections of the conformational space such that the measured dynamics can appear non-Markovian. Model-free methods to quantify FRET efficiency fluctuations would be desirable in this case. Here, we present such an approach. We determine FRET efficiency correlation functions free of artifacts from the finite length of photon trajectories or the diffusion of molecules through the confocal volume. We show that these functions capture the dynamics of proteins from nano- to milliseconds both in simulation and experiment, which provides a rigorous validation of current model-based analysis approaches.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

