Exploitable mechanisms of antibody and CAR mediated macrophage cytotoxicity
- PMID: 40595560
- PMCID: PMC12216399
- DOI: 10.1038/s41467-025-60745-x
Exploitable mechanisms of antibody and CAR mediated macrophage cytotoxicity
Abstract
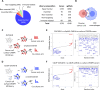
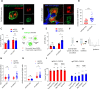
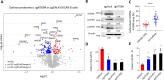
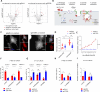
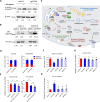
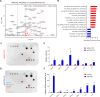
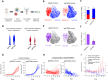
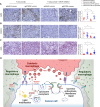
Macrophages infiltrate solid tumors and either support survival or induce cancer cell death through phagocytosis or cytotoxicity. To uncover regulators of macrophage cytotoxicity towards cancer cells, we perform two co-culture CRISPR screens using CAR-macrophages targeting different tumor associated antigens. Both identify ATG9A as an important regulator of this cytotoxic activity. In vitro and in vivo, ATG9A depletion in cancer cells sensitizes them to macrophage-mediated killing. Proteomic and lipidomic analyses reveal that ATG9A deficiency impairs the cancer cell response to macrophage-induced plasma membrane damage through defective lysosomal exocytosis, reduced ceramide production, and disrupted caveolar endocytosis. Depleting non-cytotoxic macrophages using CSF1R inhibition while preventing ATG9A-mediated tumor membrane repair enhances the anti-tumor activity of therapeutic antibodies in mice. Thus, macrophage cytotoxicity plays an important role in tumor elimination during antibody or CAR-macrophage treatment, and inhibiting tumor membrane repair via ATG9A, particularly in combination with cytotoxic macrophage enrichment through CSF1R inhibition, improves tumor-targeting macrophage efficacy.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: F.Y.F. has consulted for Astellas, Bayer, Blue Earth Diagnostics, BMS, EMD Serono, Exact Sciences, Foundation Medicine, Janssen Oncology, Myovant, Roivant, and Varian, and serves on the Scientific Advisory Board for BlueStar Genomics and SerImmune. F.Y.F. has patent applications with Decipher Biosciences, as well as with PFS Genomics/Exact Sciences in breast cancer, all unrelated to this work. L.A.G. has filed patents on CRISPR tools and CRISPR functional genomics, is a co-founder of Chroma Medicine, and a consultant for Chroma Medicine and Arena Bioworks. The remaining authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

