RFC1 regulates the expansion of neural progenitors in the developing zebrafish cerebellum
- PMID: 40595562
- PMCID: PMC12217872
- DOI: 10.1038/s41467-025-60775-5
RFC1 regulates the expansion of neural progenitors in the developing zebrafish cerebellum
Abstract
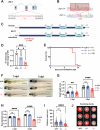
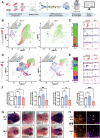
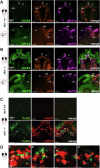
DNA replication and repair are basic yet essential molecular processes for all cells. RFC1 encodes the largest subunit of the Replication Factor C, an essential clamp-loader for DNA replication and repair. Intronic repeat expansion in RFC1 has recently been associated with so-called RFC1-related disorders, which mainly encompass late-onset cerebellar ataxias. However, the mechanisms making certain tissues more susceptible to defects in these universal pathways remain mysterious. Here, we provide the first investigation of RFC1 gene function in vivo using zebrafish. We showed that RFC1 is expressed in neural progenitor cells within the developing cerebellum, where it maintains their genomic integrity during neurogenic maturation. Accordingly, RFC1 loss-of-function leads to a severe cerebellar phenotype due to impaired neurogenesis of both Purkinje and granule cells. Our data point to a specific role of RFC1 in the developing cerebellum, paving the way for a better understanding of the pathogenic mechanisms underlying RFC1-related disorders.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.S. is a co-founder of DanioDesign Inc. (QC, Canada) and of Osta Therapeutics (France); N.P. is a co-founder of Neurenati Therapeutics (QC, Canada). These commercial affiliations did not play any role in investigational design, data collection and analysis, the decision to publish or the preparation of the manuscript. The remaining authors declare no competing interests.
Figures



References
-
- Voogd, J. The human cerebellum. J. Chem. Neuroanat.26, 243–252 (2003). - PubMed
-
- Purves, D. et al. Circuits within the Cerebellum (Sinauer Associates, 2001).
-
- Fine, E. J., Ionita, C. C. & Lohr, L. The history of the development of the cerebellar examination. Semin. Neurol.22, 375–384 (2002). - PubMed
-
- Roostaei, T., Nazeri, A., Sahraian, M. A. & Minagar, A. The human cerebellum: a review of physiologic neuroanatomy. Neurol. Clin.32, 859–869 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

