Enrichment of extracellular vesicles using Mag-Net for the analysis of the plasma proteome
- PMID: 40595564
- PMCID: PMC12219689
- DOI: 10.1038/s41467-025-60595-7
Enrichment of extracellular vesicles using Mag-Net for the analysis of the plasma proteome
Abstract
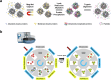
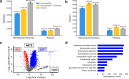
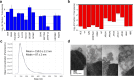
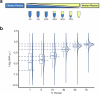
Extracellular vesicles (EVs) in plasma are composed of exosomes, microvesicles, and apoptotic bodies. We report a plasma EV enrichment strategy using magnetic beads called Mag-Net. Proteomic interrogation of this plasma EV fraction enables the detection of proteins that are beyond the dynamic range of liquid chromatography-mass spectrometry of unfractionated plasma. Mag-Net is robust, reproducible, inexpensive, and requires <100 μL plasma input. Coupled to data-independent mass spectrometry, we demonstrate the measurement of >37,000 peptides from >4,000 proteins. Using Mag-Net on a pilot cohort of patients with neurodegenerative disease and healthy controls, we find 204 proteins that differentiate (q-value < 0.05) patients with Alzheimer's disease dementia (ADD) from those without ADD. There are also 310 proteins that differ between individuals with Parkinson's disease and without. Using machine learning we distinguish between individuals with ADD and not ADD with an area under the receiver operating characteristic curve (AUROC) = 0.98 ± 0.06.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Ireshyn Govender, Stoyan Stoychev and Justin Jordaan are employed by ReSyn Biosciences, proprietors of MagReSyn® technology. The MacCoss Lab at the University of Washington (C.C.W., K.A.T., J.P., D.P., G.M., E.H., M.R., and M.J.M.) has a sponsored research agreement with Thermo Fisher Scientific, the manufacturer of the mass spectrometry instrumentation used in this research. In addition, Michael MacCoss is a paid consultant for Thermo Fisher Scientific. The remaining authors declare no competing interests.
Figures



References
-
- Anderson, N. L. & Anderson, N. G. The human plasma proteome: History, character, and diagnostic prospects*. Mol. Cell. Proteom.1, 845–867 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

