Interdependence of Pasha and Drosha for localization and function of the Microprocessor in C. elegans
- PMID: 40595566
- PMCID: PMC12219410
- DOI: 10.1038/s41467-025-60721-5
Interdependence of Pasha and Drosha for localization and function of the Microprocessor in C. elegans
Abstract
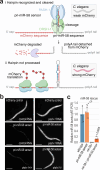
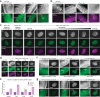
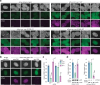
Primary microRNA (pri-miRNA) transcripts are processed by the Microprocessor, containing the ribonuclease Drosha and its RNA-binding partner DGCR8/Pasha. In a forward genetic screen utilizing a fluorescence-based sensor that monitors pri-miRNA processing in live Caenorhabditis elegans, we identify a mutation in the conserved G179 residue adjacent to the namesake W180 of Pasha's WW domain that disrupts pri-miRNA processing. We show that both the G179 and W180 residues are required for Pasha dimerization and Microprocessor assembly. The WW domain also facilitates nuclear localization of Pasha, likely through its role in Microprocessor assembly, which in turn promotes nuclear enrichment of Drosha. Furthermore, depletion of Pasha mislocalizes Drosha to the cytoplasm, and vice versa, while deletion of Pasha's N-terminus causes both proteins to accumulate in nucleoli. Our results reveal a mutual dependency between Pasha and Drosha for their localization in C. elegans and highlight the role of Pasha's WW domain in maintaining Microprocessor integrity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Regulation of Microprocessor assembly and localization via Pasha's WW domain in C. elegans.bioRxiv [Preprint]. 2024 Apr 23:2024.04.23.590772. doi: 10.1101/2024.04.23.590772. bioRxiv. 2024. PMID: 38712061 Free PMC article. Preprint.
-
Processing of primary microRNAs by the Microprocessor complex.Nature. 2004 Nov 11;432(7014):231-5. doi: 10.1038/nature03049. Epub 2004 Nov 7. Nature. 2004. PMID: 15531879
-
The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs.Mol Cell. 2024 Nov 7;84(21):4175-4190.e6. doi: 10.1016/j.molcel.2024.09.008. Epub 2024 Oct 4. Mol Cell. 2024. PMID: 39368465
-
Shared and unique mechanisms of RNAi-mediated antiviral immunity in C. elegans.Virology. 2025 Apr;605:110459. doi: 10.1016/j.virol.2025.110459. Epub 2025 Feb 21. Virology. 2025. PMID: 40022946 Review.
-
Drosha in primary microRNA processing.Cold Spring Harb Symp Quant Biol. 2006;71:51-7. doi: 10.1101/sqb.2006.71.041. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381280 Review.
References
-
- Ha, M. & Kim, V. N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014). - PubMed
-
- Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature425, 415–419 (2003). - PubMed
-
- Gregory, R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature432, 235–240 (2004). - PubMed
-
- Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the microprocessor complex. Nature432, 231–235 (2004). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

