DNA methylation protects cancer cells against senescence
- PMID: 40595593
- PMCID: PMC12216915
- DOI: 10.1038/s41467-025-61157-7
DNA methylation protects cancer cells against senescence
Abstract
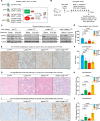
Inhibitors of DNA methylation such as 5-aza-deoxycytidine are widely used in experimental and clinical settings. However, their mechanism of action is such that DNA damage inevitably co-occurs with loss of DNA methylation, making it challenging to discern their respective effects. Here we deconvolute the effects of decreased DNA methylation and DNA damage on cancer cells, by using degron alleles of key DNA methylation regulators. We report that cancer cells with decreased DNA methylation-but no DNA damage-enter cellular senescence, with G1 arrest, SASP expression, and SA-β-gal positivity. This senescence is independent of p53 and Rb, but involves p21, which is cytoplasmic and inhibits apoptosis, and cGAS, playing a STING-independent role in the nucleus. Xenograft experiments show that tumor cells can be made senescent in vivo by decreasing DNA methylation. These findings reveal the intrinsic effects of loss of DNA methylation in cancer cells and have practical implications for future therapeutic approaches.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
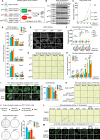
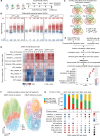
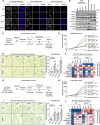
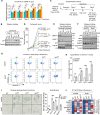

References
MeSH terms
Substances
Grants and funding
- ANR-23-CE12-0015-01/Agence Nationale de la Recherche (French National Research Agency)
- INCA_18350/Institut National Du Cancer (French National Cancer Institute)
- EQU202503019988/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- FDT202404018544/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

