The loss of OPA1 accelerates intervertebral disc degeneration and osteoarthritis in aged mice
- PMID: 40595596
- PMCID: PMC12218067
- DOI: 10.1038/s41467-025-60933-9
The loss of OPA1 accelerates intervertebral disc degeneration and osteoarthritis in aged mice
Abstract
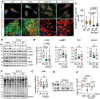
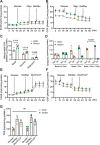
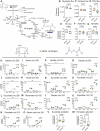
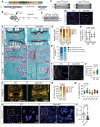
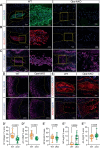
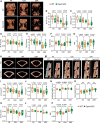
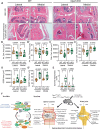
Recent studies have highlighted the importance of mitochondria in NP cells and articular chondrocyte health. Since the understanding of mechanisms governing mitochondrial dynamics in these tissues is lacking, we investigated the role of OPA1, a mitochondrial fusion protein, in their homeostasis. OPA1 knockdown in NP cells altered mitochondrial size and cristae shape and increased the oxygen consumption rate. OPA1 governed the morphology of multiple organelles, including peroxisomes, early endosomes and cis-Golgi and loss resulted in the dysregulation of autophagy. Metabolic profiling and 13C-flux analyses revealed TCA cycle anaplerosis and altered metabolism in OPA1-deficient NP cells. Noteworthy, Opa1AcanCreERT2 mice showed age-dependent disc degeneration, osteoarthritis, and vertebral osteopenia. RNA-Sequencing of Opa1cKO NP tissue revealed dysregulation of metabolism, autophagy, cytoskeletal reorganization, and extracellular matrix and shared strong thematic similarities with a subset of human degenerative NP samples. Our findings underscore that maintenance of mitochondrial dynamics and multi-organelle cross-talk is critical in preserving metabolic homeostasis of disc and cartilage.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Update of
-
The loss of OPA1 accelerates intervertebral disc degeneration and osteoarthritis in aged mice.Res Sq [Preprint]. 2024 Feb 20:rs.3.rs-3950044. doi: 10.21203/rs.3.rs-3950044/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Jul 1;16(1):5996. doi: 10.1038/s41467-025-60933-9. PMID: 38464287 Free PMC article. Updated. Preprint.
References
-
- Disease, G. B. D., Injury, I. & Prevalence, C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet392, 1789–1858 (2018). - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01 AR074813/AR/NIAMS NIH HHS/United States
- R01AR055655/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- R01 AG078609/AG/NIA NIH HHS/United States
- R35 GM144103/GM/NIGMS NIH HHS/United States
- R01AR074813/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

