Binge drinking triggers VGLUT3-mediated glutamate secretion and subsequent hepatic inflammation by activating mGluR5/NOX2 in Kupffer cells
- PMID: 40595616
- PMCID: PMC12216207
- DOI: 10.1038/s41467-025-60820-3
Binge drinking triggers VGLUT3-mediated glutamate secretion and subsequent hepatic inflammation by activating mGluR5/NOX2 in Kupffer cells
Abstract
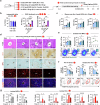
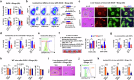
Glutamate, a crucial player in hepatic amino acid metabolism, has been relatively unexplored in immune cell activation. We show in a study with male mice that hepatic glutamate accumulates in vesicles of perivenous hepatocytes through vesicular glutamate transporter 3 (VGLUT3), regulated by the aryl hydrocarbon receptor upon chronic alcohol intake. Additional binge drinking triggers the exocytosis of glutamate by altering the intracellular Ca2+ level, stimulating metabotropic glutamate receptor 5 (mGluR5) and subsequent NADPH oxidase 2 (NOX2)-mediated ROS production in Kupffer cells (KCs). This interaction between hepatocytes and KCs is facilitated by pseudosynapse formation, arising from alcohol-induced ballooning of perivenous hepatocytes. Genetic or pharmacological interference of mGluR5 or NOX2 in KCs alleviates alcohol-related steatohepatitis (ASH). Analysis of patient samples confirmed some of the findings from mice, showing that plasma glutamate concentration and VGLUT3 levels correlate with ASH development. Conclusively, our findings highlight glutamate storage and release in mediating ASH, particularly through the pseudosynapse between hepatocytes and KCs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
HIF-2α drives hepatic Kupffer cell death and proinflammatory recruited macrophage activation in nonalcoholic steatohepatitis.Sci Transl Med. 2024 Sep 11;16(764):eadi0284. doi: 10.1126/scitranslmed.adi0284. Epub 2024 Sep 11. Sci Transl Med. 2024. PMID: 39259813 Free PMC article.
-
TLR8 agonist selgantolimod regulates Kupffer cell differentiation status and impairs HBV entry into hepatocytes via an IL-6-dependent mechanism.Gut. 2024 Nov 11;73(12):2012-2022. doi: 10.1136/gutjnl-2023-331396. Gut. 2024. PMID: 38697771 Free PMC article.
-
Kupffer cell programming by maternal obesity triggers fatty liver disease.Nature. 2025 Aug;644(8077):790-798. doi: 10.1038/s41586-025-09190-w. Epub 2025 Jun 18. Nature. 2025. PMID: 40533564 Free PMC article.
-
Selenium for preventing cancer.Cochrane Database Syst Rev. 2018 Jan 29;1(1):CD005195. doi: 10.1002/14651858.CD005195.pub4. Cochrane Database Syst Rev. 2018. PMID: 29376219 Free PMC article.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
References
-
- Seitz, H. K. et al. Alcoholic liver disease. Nat. Rev. Dis. Prim.4, 16 (2018). - PubMed
-
- Singal, A. K. & Mathurin, P. Diagnosis and treatment of alcohol-associated liver disease: a review. JAMA326, 165–176 (2021). - PubMed
-
- Lucey, M. R., Mathurin, P. & Morgan, T. R. Alcoholic hepatitis. N. Engl. J. Med.360, 2758–2769 (2009). - PubMed
-
- Krenkel, O. & Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol.17, 306–321 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

