Bridging cell morphological behaviors and molecular dynamics in multi-modal spatial omics with MorphLink
- PMID: 40595621
- PMCID: PMC12217155
- DOI: 10.1038/s41467-025-61142-0
Bridging cell morphological behaviors and molecular dynamics in multi-modal spatial omics with MorphLink
Abstract
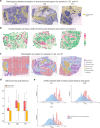
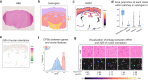
Multi-modal spatial omics data are invaluable for exploring complex cellular behaviors in diseases from both morphological and molecular perspectives. Current analytical methods primarily focus on clustering and classification, and do not adequately examine the relationship between cell morphology and molecular dynamics. Here, we present MorphLink, a framework designed to systematically identify disease-related morphological-molecular interplays. MorphLink has been evaluated across a wide array of datasets, showcasing its effectiveness in extracting and linking interpretable morphological features with various molecular measurements in spatial omics analyses. These linkages provide a transparent view of cellular behavior heterogeneity within tissue regions with similar cell type compositions, characterizing tumor subtypes and immune diversity across different organs. Additionally, MorphLink is scalable and robust against cross-sample batch effects, making it an efficient method for integrative spatial omics data analysis across samples, cohorts, and modalities, and enhancing the interpretation of results for large-scale studies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: This study was supported by start-up research funds from the Department of Human Genetics, School of Medicine at Emory University. The authors declare no competing interests.
Figures




Update of
-
MorphLink: Bridging Cell Morphological Behaviors and Molecular Dynamics in Multi-modal Spatial Omics.bioRxiv [Preprint]. 2024 Aug 26:2024.08.24.609528. doi: 10.1101/2024.08.24.609528. bioRxiv. 2024. Update in: Nat Commun. 2025 Jul 1;16(1):5878. doi: 10.1038/s41467-025-61142-0. PMID: 39253421 Free PMC article. Updated. Preprint.
References
-
- Fitzgibbons, P. L. et al. Prognostic factors in breast cancer: College of American Pathologists consensus statement 1999. Arch. Pathol. Lab. Med.124, 966–978 (2000). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

