Human neutrophil α-defensin HNP1 interacts with bacterial OmpA to promote Acinetobacter baumannii biofilm formation
- PMID: 40595622
- PMCID: PMC12216189
- DOI: 10.1038/s41467-025-60935-7
Human neutrophil α-defensin HNP1 interacts with bacterial OmpA to promote Acinetobacter baumannii biofilm formation
Abstract
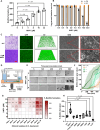
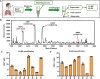
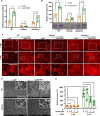
Acinetobacter baumannii is the causative agent of a wide range of nosocomial and community-acquired infections that remain extremely difficult to treat due largely to its antibiotic resistance contributed, in part, by biofilm formation. We find that the prototypic human neutrophil α-defensin HNP1, present in the bronchoalveolar lavage fluids from Acinetobacter baumannii-infected patients, promotes Acinetobacter baumannii biofilm formation through interactions with the bacterial outer membrane protein OmpA. As a result of HNP1-enhanced biofilm formation, Acinetobacter baumannii becomes more tolerant to antibiotics and more readily colonizes host cells and tissues. These unexpected findings contrast the protective roles HNP1 plays in innate immunity against microbial infection, showcasing an example of the host-pathogen arms race where a host defense peptide is exploited by a microbe for pathogenicity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Relationship between antibiotic resistance and biofilm-forming capacity with RND efflux pumps expression in clinical Acinetobacter baumannii isolates.Arch Microbiol. 2025 May 25;207(7):153. doi: 10.1007/s00203-025-04347-7. Arch Microbiol. 2025. PMID: 40413665
-
Characterizing the interplay between Acinetobacter baumannii, A549 cells, and anti-Omp34 antibodies: implications for adherence, internalization, and cytotoxicity.Folia Microbiol (Praha). 2025 Jun;70(3):699-709. doi: 10.1007/s12223-024-01218-4. Epub 2024 Oct 31. Folia Microbiol (Praha). 2025. PMID: 39480642
-
In silico and in vitro comparative analysis of 79 Acinetobacter baumannii clinical isolates.Microbiol Spectr. 2025 Jul;13(7):e0284924. doi: 10.1128/spectrum.02849-24. Epub 2025 May 16. Microbiol Spectr. 2025. PMID: 40377313 Free PMC article.
-
Unveiling the role of adhesin proteins in controlling Acinetobacter baumannii infections: a systematic review.Infect Immun. 2025 Feb 18;93(2):e0034824. doi: 10.1128/iai.00348-24. Epub 2025 Jan 8. Infect Immun. 2025. PMID: 39772848 Free PMC article.
-
Post-neurosurgical multidrug-resistant Acinetobacter baumannii meningitis successfully treated with intrathecal colistin. A new case and a systematic review of the literature.Int J Infect Dis. 2010 Jul;14(7):e572-9. doi: 10.1016/j.ijid.2009.06.032. Epub 2009 Nov 4. Int J Infect Dis. 2010. PMID: 19892577
References
-
- Whiteway, C., Breine, A., Philippe, C. & Van der Henst, C. Acinetobacter baumannii. Trends Microbiol.30, 199–200 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- 82030062/National Natural Science Foundation of China (National Science Foundation of China)
- 82202529/National Natural Science Foundation of China (National Science Foundation of China)
- ZD2021CY001/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
- 2021M690666/China Postdoctoral Science Foundation
- 2022T150129/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Research Materials

