Adhesin Als4112 promotes Candida auris skin colonization through interactions with keratinocytes and extracellular matrix proteins
- PMID: 40595627
- PMCID: PMC12214902
- DOI: 10.1038/s41467-025-60876-1
Adhesin Als4112 promotes Candida auris skin colonization through interactions with keratinocytes and extracellular matrix proteins
Abstract
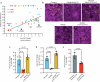
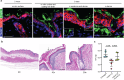
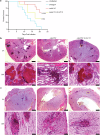
Candida auris is a fungal pathogen notorious for persistent skin colonization and transmission in healthcare settings. Here, we show that a C. auris conserved adhesin, Als4112, is required for skin colonization via keratinocyte attachment and direct interactions with host extracellular matrix proteins, especially basement membrane proteins such as laminin. Deletion of ALS4112 reduces skin colonization in mouse models of epicutaneous and systemic infection. In addition, coating plastic and catheter surfaces with collagen I or III inhibits C. auris attachment and biofilm formation. Our study highlights the critical role of Als4112 in C. auris colonization and virulence, and explores potential strategies to reduce the pathogen's adherence to abiotic surfaces and thus its spread in healthcare settings.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
In vitro and in vivo activity of sodium houttuyfonate and sodium new houttuyfonate against Candida auris infection by affecting adhesion, aggregation, and biofilm formation abilities.Microbiol Spectr. 2025 Aug 5;13(8):e0022225. doi: 10.1128/spectrum.00222-25. Epub 2025 Jun 18. Microbiol Spectr. 2025. PMID: 40530673 Free PMC article.
-
A Candida auris-specific adhesin, Scf1, governs surface association, colonization, and virulence.Science. 2023 Sep 29;381(6665):1461-1467. doi: 10.1126/science.adf8972. Epub 2023 Sep 28. Science. 2023. PMID: 37769084 Free PMC article.
-
The calcineurin pathway regulates extreme thermotolerance, cell membrane and wall integrity, antifungal resistance, and virulence in Candida auris.PLoS Pathog. 2025 Jul 28;21(7):e1013363. doi: 10.1371/journal.ppat.1013363. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40720551 Free PMC article.
-
Key Factors to Consider for Candida auris Screening in Healthcare Settings: A Systematic Review.Mycoses. 2025 Mar;68(3):e70043. doi: 10.1111/myc.70043. Mycoses. 2025. PMID: 40072118
-
Head skin infection by Candida auris: A case report.J Mycol Med. 2025 Jun;35(2):101544. doi: 10.1016/j.mycmed.2025.101544. Epub 2025 Mar 18. J Mycol Med. 2025. PMID: 40147064 Review.
Cited by
-
Candida auris Metabolism and Growth Preferences in Physiologically Relevant Skin-like Conditions.bioRxiv [Preprint]. 2025 May 19:2025.05.18.654780. doi: 10.1101/2025.05.18.654780. bioRxiv. 2025. PMID: 40475518 Free PMC article. Preprint.
-
Growth phase influences virulence in Candida auris systemic infection models.bioRxiv [Preprint]. 2025 Jun 2:2025.06.02.657403. doi: 10.1101/2025.06.02.657403. bioRxiv. 2025. PMID: 40501926 Free PMC article. Preprint.
References
-
- Satoh, K. et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol.53, 41–44 (2009). - PubMed
-
- Mulet Bayona, J. V., Tormo Palop, N., Salvador García, C., Guna Serrano, M. D. R. & Gimeno Cardona, C. Candida auris from colonisation to candidemia: A four-year study. Mycoses66, 882–890 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

