Proteomic analysis of Down syndrome cerebrospinal fluid compared to late-onset and autosomal dominant Alzheimer´s disease
- PMID: 40595720
- PMCID: PMC12214755
- DOI: 10.1038/s41467-025-61054-z
Proteomic analysis of Down syndrome cerebrospinal fluid compared to late-onset and autosomal dominant Alzheimer´s disease
Abstract
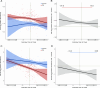
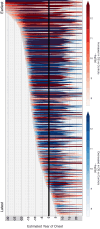
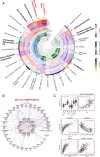
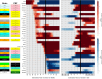
Almost all individuals with Down Syndrome (DS) develop Alzheimer's disease (AD) by mid to late life. However, the degree to which AD in DS shares pathological changes with sporadic late-onset AD (LOAD) and autosomal dominant AD (ADAD) beyond core AD biomarkers such as amyloid-β (Aβ) and tau is unknown. Here, we used proteomics of cerebrospinal fluid from individuals with DS (n = 229) in the Down Alzheimer Barcelona Neuroimaging Initiative (DABNI) cohort to assess the evolution of AD pathophysiology from asymptomatic to dementia stages and compared the proteomic biomarker changes in DS to those observed in LOAD and ADAD. Although many proteomic alterations were shared across DS, LOAD, and ADAD, DS demonstrated more severe changes in immune-related proteins, extracellular matrix pathways, and plasma proteins likely related to blood-brain barrier dysfunction compared to LOAD. These changes were present in young adults with DS prior to the onset of Aβ or tau pathology, suggesting they are associated with trisomy 21 and may serve as risk factors for DSAD. DSAD showed an earlier increase in markers of axonal and white matter pathology and earlier changes in markers potentially associated with cerebral amyloid angiopathy compared to ADAD. The unique features of DSAD may have important implications for treatment strategies in this population.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.C.B.J. has served on an advisory board for Eli Lilly and has received royalties from EmTheraPro (outside submitted work). J.F. reported receiving personal fees for service on the advisory boards, adjudication committees or speaker honoraria from AC Immune, Adamed, Alzheon, Biogen, Eisai, Esteve, Fujirebio, Ionis, Laboratorios Carnot, Life Molecular Imaging, Lilly, Lundbeck, Perha, Roche and outside the submitted work. O.B., D.A., A.L., and J.F. report holding a patent for markers of synaptopathy in neurodegenerative disease (WO2019175379, applicant Fundació Institut de Recerca de L’hospital de la Santa Creu, Sant Pau, “Markers of synaptopathy in neurodegenerative diseases”, EP19709749.6, priority date 16/3/2018, patent pending, inventors O Belbin, A Lleó, A Bayés, J Fortea, and D Alcolea, licensed to ADX NeuroSciences, N.V. EPI8382175.0 to develop antibodies and immunoassays for Calsyntenin-1, GluR2, GluR4, Neuroligin-2, neurexin-2a, neurexin-3a, syntaxin-1b, thy-1, tenascin-r, and vamp-2). N.T.S. and A.I.L. are founders of EmTheraPro (outside submitted work). H.Z. has served at scientific advisory boards and/or as a consultant for Abbvie, Acumen, Alector, Alzinova, ALZPath, Annexon, Apellis, Artery Therapeutics, AZTherapies, Cognito Therapeutics, CogRx, Denali, Eisai, Nervgen, Novo Nordisk, Optoceutics, Passage Bio, Pinteon Therapeutics, Prothena, Red Abbey Labs, reMYND, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure, Biogen, and Roche, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). K.B. has served as a consultant, on advisory boards, or at data monitoring committees for Abcam, Axon, BioArctic, Biogen, and JOMDD/Shimadzu. Julius Clinical, Lilly, MagQu, Novartis, Ono Pharma, Pharmatrophix, Prothena, Roche Diagnostics, and Siemens Healthineers, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program, outside the work presented in this paper. T.W. has served as a consultant or on data monitoring committees for Acumen, Biogen, Grifols, Lilly and ProMIS Neurosciences. The remaining authors declare no competing interests.
Figures

References
-
- McCarron, M. et al. A prospective 20-year longitudinal follow-up of dementia in persons with Down syndrome. J. Intellect. Disabil. Res.61, 843–852 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

