Unveiling diagnostic biomarkers and therapeutic targets in lung adenocarcinoma using bioinformatics and experimental validation
- PMID: 40595823
- PMCID: PMC12218858
- DOI: 10.1038/s41598-025-05227-2
Unveiling diagnostic biomarkers and therapeutic targets in lung adenocarcinoma using bioinformatics and experimental validation
Abstract
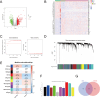
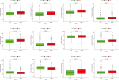
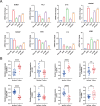
Lung adenocarcinoma (LUAD) is a major challenge in oncology due to its complex molecular structure and generally poor prognosis. The aim of this study was to find diagnostic markers and therapeutic targets for LUAD by integrating differential gene expression analysis, weighted gene co-expression network analysis (WGCNA), and machine learning methods. Differentially expressed genes (DEGs) were identified through the analysis of gene expression data from the Gene Expression Omnibus (GEO) database. Ten of the gene co-expression modules constructed by WGCNA were identified, with the red module having the most significant correlation with clinical features. In addition, a machine learning model constructed based on Stepglm[backward] with the random forest algorithm achieved the highest C-index (0.999) and screened eight core genes, among which ST14 was noted for its excellent predictive ability. Single-cell RNA sequencing of the LUAD samples further analyzed the expression patterns of these genes in 29 cellular subtypes, revealing their significant association with immune cell infiltration. Of particular note, the association of ST14 with clinical prognosis, drug responsiveness, and immune infiltration was validated, while enrichment analysis further clarified its role in key biological pathways. Ultimately, the expression of the core genes was validated experimentally. This study provides new insights into the pathogenesis of LUAD, clarifies potential diagnostic markers and therapeutic targets, and provides an important basis for future clinical interventions.
Keywords: Lung adenocarcinoma; Machine learning in oncology; ST14; Single-cell analysis; Weighted gene co-expression network analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments. The project was approved by the institutional ethics committee of the Fujian Cancer Hospital. Informed consent was obtained from all participants included in the study. Competing interests: The authors declare no competing interests.
Figures





References
-
- Duma, N., Santana-Davila, R. & Molina, J. R. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc.94, 1623–1640. 10.1016/j.mayocp.2019.01.013 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

