Genomic characterisation of nasal isolates of coagulase-negative Staphylococci from healthy medical students reveals novel Staphylococcal cassette chromosome mec elements
- PMID: 40595841
- PMCID: PMC12216672
- DOI: 10.1038/s41598-025-05159-x
Genomic characterisation of nasal isolates of coagulase-negative Staphylococci from healthy medical students reveals novel Staphylococcal cassette chromosome mec elements
Abstract
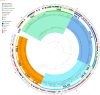
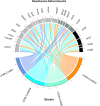
Coagulase-negative staphylococci (CoNS) are a diverse group of Gram-positive bacteria that are part of the normal human microbiota. Once thought to be non-pathogenic, CoNS has emerged in recent years as opportunistic pathogens of concern particularly in healthcare settings. In this study, the genomes of four methicillin-resistant CoNS isolates obtained from the nasal swabs of healthy university medical students in Malaysia were sequenced using the Illumina short-read platform. Genome sequencing enabled the identification of the four isolates as Staphylococcus warneri UTAR-CoNS1, Staphylococcus cohnii subsp. cohnii UTAR-CoNS6, Staphylococcus capitis subsp. urealyticus UTAR-CoNS20, and Staphylococcus haemolyticus UTAR-CoNS26. The genome of S. cohnnii UTAR-CoNS6 harboured the mecA methicillin-resistance gene on a Staphylococcal cassette chromosome mec (SCCmec) element similar to SCCmec type XIV (5 A) but the SCCmec cassettes identified in the other three CoNS genomes were novel and untypeable. Some of these SCCmec elements also encoded heavy metal resistance genes while the SCCmec type XIV (5 A) variant in S. cohnii UTAR-CoNS6 harboured the complete ica operon, a known virulence factor that functions in biofilm formation. In S. cohnii UTAR-CoNS6, the macrolide resistance genes msrA and mphC along with copper and cadmium resistance genes were located on a 26,630 bp plasmid, pUCNS6. This study showcased the diversity of CoNS in the nasal microbiota of medical students but the discovery of novel SCCmec elements, various antimicrobial and heavy metal resistance along with virulence genes in these isolates is of concern and warrants vigilance due to the likelihood of spread, especially to hospitalised patients.
Keywords: Coagulase-negative Staphylococci (CoNS); Plasmids; SCCmec; Whole-genome sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- WHO. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. https://www.who.int/publications/i/item/9789240093461 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

