Identification of a LncRNA based CeRNA network signature to establish a prognostic model and explore potential therapeutic targets in gastric cancer
- PMID: 40595924
- PMCID: PMC12219596
- DOI: 10.1038/s41598-025-05105-x
Identification of a LncRNA based CeRNA network signature to establish a prognostic model and explore potential therapeutic targets in gastric cancer
Abstract
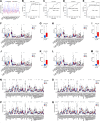
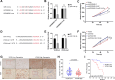
Numerous studies have demonstrated that long non-coding RNA (lncRNA) play critical roles in regulating physiological processes and contributing to pathological diseases. This study aimed to develop lncRNA-based signatures to predict the prognostic risk of gastric cancer (GC) patients and provide therapeutic guidance. Gene expression profiles and clinical information were obtained from The Cancer Genome Atlas (TCGA) database. Differentially expressed RNAs, including lncRNA, miRNA, and mRNA, in cancerous and adjacent non-cancerous tissues were analyzed using Weighted correlation network analysis (WGCNA) and construction of a lncRNA-miRNA-mRNA competing endogenous RNA (ceRNA) network. Then, a lncRNA-based risk model was constructed by Cox regression and Lasso regression analyses. A ceRNA network comprising 235 lncRNAs, 60 miRNAs, and 52 mRNAs was identified. Based on the expression of five lncRNAs (including AC010333.1, LINC01579, AP000695.2, LINC00922 and AL121772.1) screened from the ceRNA network, a lncRNA-based risk model was developed, which effectively predict the prognosis of GC patients. The expression of AP000695.2 was significantly associated with poor prognosis and higher T stage. The knockdown of AP000695.2 inhibited the growth of GC cells both in vitro and in vivo. Transfection with miR-144-3p and miR-7-5p mimics attenuate the up-regulation of targets genes, including CDH11, COL5A2, COL12A1, and VCAN, which was induced by AP000695.2, suggesting a ceRNA mechanism. Additionally, elevated VCAN expression was correlated with poorer survival and a reduced response to anti-PD-1 immune checkpoint inhibitor treatment of GC. This study established a lncRNA-based risk model for predicting the prognosis of GC patients and identified a ceRNA mechanism involving AP000695.2-miR-144-3p-VCAN, presenting novel biomarkers and therapeutic targets for GC treatment.
Keywords: Gastric cancer; Immunotherapy; LncRNA; Prognostic model; Tumor microenvironment; WGCNA.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Genome-wide analysis of prognostic-related lncRNAs, miRNAs and mRNAs forming a competing endogenous RNA network in lung squamous cell carcinoma.J Cancer Res Clin Oncol. 2020 Jul;146(7):1711-1723. doi: 10.1007/s00432-020-03224-8. Epub 2020 Apr 30. J Cancer Res Clin Oncol. 2020. PMID: 32356177 Free PMC article.
-
Construction of an immune-related prognostic signature and lncRNA-miRNA-mRNA ceRNA network in acute myeloid leukemia.J Leukoc Biol. 2024 Jun 28;116(1):146-165. doi: 10.1093/jleuko/qiae041. J Leukoc Biol. 2024. PMID: 38393298
-
A novel copper-induced cell death-related lncRNA prognostic signature associated with immune infiltration and clinical value in gastric cancer.J Cancer Res Clin Oncol. 2023 Sep;149(12):10543-10559. doi: 10.1007/s00432-023-04916-7. Epub 2023 Jun 8. J Cancer Res Clin Oncol. 2023. PMID: 37291405 Free PMC article.
-
Multifaceted interactions between lncRNA-associated ceRNA networks and small molecules in triple-negative breast cancer.Biomed Pharmacother. 2025 Aug;189:118241. doi: 10.1016/j.biopha.2025.118241. Epub 2025 Jun 16. Biomed Pharmacother. 2025. PMID: 40527037 Review.
-
Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis.Mol Cancer. 2016 Jun 28;15(1):50. doi: 10.1186/s12943-016-0535-1. Mol Cancer. 2016. PMID: 27352941 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

