Metabolic features of patients with repeated Omicron infections highlight new targets for therapeutic intervention
- PMID: 40595933
- PMCID: PMC12216247
- DOI: 10.1038/s41598-025-04745-3
Metabolic features of patients with repeated Omicron infections highlight new targets for therapeutic intervention
Abstract
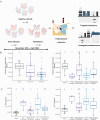
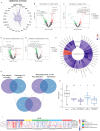
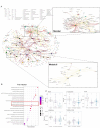
Emerging evidence underscores the role of metabolites in immunomodulation. We surmise that specific metabolic signatures might be conserved during repeated Omicron infections. To verify our hypothesis, patients with first (n = 28) and repeated Omicron infections (n = 38) between November 2023 to April 2024 were recruited into this study. Healthy controls (n = 20) were enrolled in the same period. Comprehensive serum metabolome and lipidome were quantitated using mass spectrometric approaches. The neutralizing activity of sera against the pseudotyped Omicron variant JN.1 was determined. Circulating cytokines/chemokines were quantified using a Bioplex Kit Assay. The proportion of severe/moderate infections was 2.9-fold higher in first infection patients compared to reinfection patients (67.9% vs. 23.7%, p = 0.004). Geometric mean titers (GMT) for the Omicron variant JN.1 were higher in moderate/severe infections than mild infections, but non-significant between first and repeated infections. We observed perturbed coregulation between plasma indoles and circulating plasmalogen phospholipids in Omicron-infected patients, while disrupted histidine-triacylglycerol coregulation was specific to first-infections. A panel of three lasso-selected metabolites (SL d18:1/22:0 h, tetra-peptide Pro Tyr Tyr Val, and 1,2,3,4-Tetrahydroisoquinoline) effectively differentiated moderate/severe Omicron infections from mild ones (AUROC at 0.917, 95% CI 0.793-1.000). Our findings highlight modifiable metabolic signatures as possibly new therapeutic interventions against rapidly evolving variants of SARS-CoV-2.
Keywords: Host-microbe interactions; Metabolites; Omicron; Reinfection; Severity.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
SARS-CoV-2-neutralising monoclonal antibodies to prevent COVID-19.Cochrane Database Syst Rev. 2022 Jun 17;6(6):CD014945. doi: 10.1002/14651858.CD014945.pub2. Cochrane Database Syst Rev. 2022. PMID: 35713300 Free PMC article.
-
Nirmatrelvir combined with ritonavir for preventing and treating COVID-19.Cochrane Database Syst Rev. 2023 Nov 30;11(11):CD015395. doi: 10.1002/14651858.CD015395.pub3. Cochrane Database Syst Rev. 2023. PMID: 38032024 Free PMC article.
-
Dynamics of neutralizing antibodies against COVID-19 Omicron subvariants following breakthrough infection in southwest China between December 2022 and April 2024.Signal Transduct Target Ther. 2025 Jul 30;10(1):242. doi: 10.1038/s41392-025-02319-3. Signal Transduct Target Ther. 2025. PMID: 40738879 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Neutralizing Antibody and T-Cell Spike Targeted Responses Following Receipt of a Monovalent Omicron JN.1-Adapted mRNA COVID-19 Vaccine in Immunosuppressed and Healthy Individuals.J Med Virol. 2025 Jun;97(6):e70463. doi: 10.1002/jmv.70463. J Med Virol. 2025. PMID: 40545672
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

