Unveiling how mitotic spindle orientation in 3D human colon organoids affects matrix displacements through a 4D study using DVC
- PMID: 40595987
- PMCID: PMC12215373
- DOI: 10.1038/s41598-025-04156-4
Unveiling how mitotic spindle orientation in 3D human colon organoids affects matrix displacements through a 4D study using DVC
Abstract
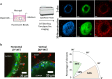
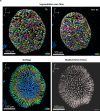
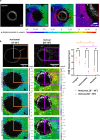
Cell division is a major event in tissue homeostasis, enabling renewal and regeneration. In human colon, vertical division is mainly observed in the stem cell compartment while horizontal division is more frequent in the progenitor transit amplifying zone. To study cell division, the human colon epithelium represents a relevant model due to its rapid renewal and high number of mitoses. Studying live mechanical interactions between the epithelium and its matrix in vivo is challenging due to the lack of suitable methods. Colon organoids seeded in Matrigel are good models because they recapitulate the organization and properties of tissue architecture. This culture set-up allows to study the displacements of the matrix around the organoid. We studied the impact of cell division within the human colonic epithelium on the extracellular matrix. We validated an original experimental and analytical process with 3D time-lapse confocal microscopy to follow cell division and matrix displacements, on which we performed a 4D Digital Volume Correlation. Depending on the orientation of the mitotic spindle, cell division affects the matrix differently. Vertical division causes a predominantly uniaxial displacement of the matrix, while horizontal division involves a multiaxial and wider displacement.
Keywords: 4D digital volume correlation; 4D displacements; Cell division; Extracellular matrix; Organoid.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Parents' and informal caregivers' views and experiences of communication about routine childhood vaccination: a synthesis of qualitative evidence.Cochrane Database Syst Rev. 2017 Feb 7;2(2):CD011787. doi: 10.1002/14651858.CD011787.pub2. Cochrane Database Syst Rev. 2017. PMID: 28169420 Free PMC article.
-
Human colonic organoids for understanding early events of familial adenomatous polyposis pathogenesis.J Pathol. 2025 Jan;265(1):26-40. doi: 10.1002/path.6366. Epub 2024 Dec 6. J Pathol. 2025. PMID: 39641466 Free PMC article.
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Does Augmenting Irradiated Autografts With Free Vascularized Fibula Graft in Patients With Bone Loss From a Malignant Tumor Achieve Union, Function, and Complication Rate Comparably to Patients Without Bone Loss and Augmentation When Reconstructing Intercalary Resections in the Lower Extremity?Clin Orthop Relat Res. 2025 Jun 26. doi: 10.1097/CORR.0000000000003599. Online ahead of print. Clin Orthop Relat Res. 2025. PMID: 40569278
References
MeSH terms
Grants and funding
- RPH21004BBA/Institut National de la Santé et de la Recherche Médicale
- RPH21004BBA/Institut National de la Santé et de la Recherche Médicale
- R21104BB/Région Occitanie Pyrénées-Méditerranée
- R21104BB/Région Occitanie Pyrénées-Méditerranée
- R21104BB/Région Occitanie Pyrénées-Méditerranée
- C20048BS/Plan Cancer MIC
- C20048BS/Plan Cancer MIC
- C20048BS/Plan Cancer MIC
- C20048BS/Plan Cancer MIC
- C18006BS/Plan Cancer 'Biology des systèmes'
- C18006BS/Plan Cancer 'Biology des systèmes'
- C18006BS/Plan Cancer 'Biology des systèmes'
- R21171BB/Agence Nationale de la Recherche
- R21171BB/Agence Nationale de la Recherche
- R21171BB/Agence Nationale de la Recherche
LinkOut - more resources
Full Text Sources

