Genomic architecture of purebred and crossbred Moghani lambs with Texel and Booroola sheep
- PMID: 40596029
- PMCID: PMC12217871
- DOI: 10.1038/s41598-025-06152-0
Genomic architecture of purebred and crossbred Moghani lambs with Texel and Booroola sheep
Abstract
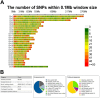
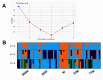
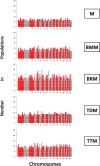
Crossbreeding with Booroola or Texel sheep harboring major genes for prolificacy and muscularity enhances productivity but it may limit adaptation and survivorship in crossbred lambs. Thus, the trade-offs that impact productivity and adaptability have not been quantified or modeled and remain largely elusive at the genetic level, limiting the development of optimized breeding strategies. This study investigates the genomic architecture of purebred Moghani sheep and the first paternal backcross (PBC1) generation of crossbred lambs, including Booroola Merino × Moghani (BMM), Booroola Romney × Moghani (BRM), Texel Dalzell × Moghani (TDM), and Texel Tamlet × Moghani (TTM). Genotyping-by-sequencing (GBS) was used to assess genetic diversity, admixture patterns, and selection signatures. Structure analysis revealed complex admixture in BMM and BRM, while TDM and TTM were more homogeneous. Purebred Moghani sheep exhibited the highest genetic diversity (HO = 0.521 ± 0.10) and the lowest inbreeding (FIS = - 0.474), serving as a key genetic bridge among the groups. In contrast, BRM and TTM showed lower heterozygosity (HO = 0.410 ± 0.09 and 0.431 ± 0.10) and increased inbreeding (FROH), with extended runs of homozygosity (ROH), suggesting recent inbreeding and reduced effective population sizes. The functional annotation of ROH islands connected TDM and TTM to immune response and muscle development pathways like VEGF and insulin signaling, while BMM and BRM were linked to metabolic and reproductive pathways like central carbon metabolism in cancer (mTOR) and prolactin signaling (LHB). Taken together, these results highlight the need for improved breeding methods that prioritize trade-offs associated with reduced genetic diversity in crossbred populations. Nevertheless, given the limited genotype representation, the results should be taken with caution; so, next research should cover a larger panel of genotypes in order to have a more complete knowledge.
Keywords: Genetic diversity; Genomic architecture; Genotyping-by-sequencing; Inbreeding; Sheep.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures







References
-
- World Livestock 2011—Livestock in food security. http://www.fao.org/3/i2373e/i2373e00.htm.
-
- Talebi, R. et al. Detection of single nucleotide polymorphisms at major prolificacy genes in the Mehraban sheep and association with litter size. Ann. Anim. Sci.18, 685 (2018).
-
- Talebi, R. et al. Association of ovine follistatin gene polymorphisms with body measurements, fat-tail traits and morphometric of head in Iranian Mehraban sheep. Small Rumin. Res.225, 107020 (2023).
-
- Majd, S. A. et al. Polymorphism identification in ovine KISS1R/GPR54 gene among pure and crossbreeds of Iranian sheep. Small Rumin. Res.173, 23–29 (2019).
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

