A single-cell atlas of the murine limb skeleton integrating the developmental and adult stages
- PMID: 40596066
- PMCID: PMC12215971
- DOI: 10.1038/s41598-025-05277-6
A single-cell atlas of the murine limb skeleton integrating the developmental and adult stages
Abstract
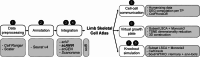
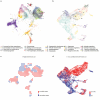
The recent growth of single-cell transcriptomics has made single-cell RNA sequencing (scRNA-seq) into a near-routine technique. Breakthroughs in scalability have led to the creation of organism-wide transcriptomic datasets, aiming to comprehensively profile the cell types and states within an organism throughout its lifecycle. However, the skeleton remains an underrepresented organ system in organism-wide atlases. Given the skeleton's critical role as the central framework of the vertebrate body, its function in housing the hematopoietic niche, and its involvement in metabolic and homeostatic processes, its underrepresentation presents a significant gap in current reference atlas projects. To address this issue, we integrated ten separate murine, publicly available scRNA-seq datasets, which include limb skeletal cells and their developmental precursors, resulting in an atlas of 133,332 cells. This limb skeletal cell atlas describes cells within the mesenchymal lineage, focusing on the process from limb induction to adult bone formation, and encompasses 39 well-characterized cell types and states. By expanding the repertoire of time points and cell types within a single dataset, we enable more complete analyses of cell-cell communication or in silico perturbation studies. Together, these efforts present a valuable resource for researchers in skeletal biology, metabolism, and regenerative medicine, filling an important gap in current atlas mapping projects.
Keywords: Atlas; Bioinformatics; Bone; Limb; Morphogenesis; Single-cell; Transcriptome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
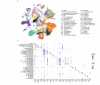


References
-
- Einhorn, T. A. The cell and molecular biology of fracture healing. Clin. Orthop. Relat. Res.355, S7−21. 10.1097/00003086-199810001-00003 (1998). - PubMed
-
- Asp, M. et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 179, 1647−1660. 10.1016/j.cell.2019.11.025 (2019). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

