Revealing the composition of bacterial communities in various oil-contaminated soils and investigating their intrinsic traits in hydrocarbon degradation
- PMID: 40596159
- PMCID: PMC12219062
- DOI: 10.1038/s41598-025-05519-7
Revealing the composition of bacterial communities in various oil-contaminated soils and investigating their intrinsic traits in hydrocarbon degradation
Abstract
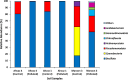
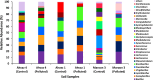
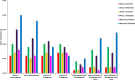
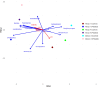
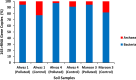
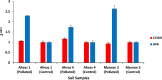
This study explores prokaryotic diversity and oil biodegradation potential in soils from three evaporation ponds in the Ahvaz and Maroon oil fields, Iran. Despite prior studies on prokaryotic diversity in contaminated soils, systematic comparisons within the same region remain limited. The analysis identified distinct physicochemical differences across sites. Ahvaz 1 soil, with a loamy silty clay texture, had the highest salinity (15.4%) and total petroleum hydrocarbons (TPH, 3.5%). Ahvaz 4 soil, loamy silty in texture, showed 7.49% salinity and 1% TPH, while Maroon 3 soil exhibited the lowest salinity (5.06%) and TPH (0.5%). Prokaryotic diversity and biodegradation traits were assessed using 16S rRNA next-generation sequencing (NGS) and qPCR, respectively. NGS revealed reduced prokaryotic diversity in all contaminated soils, with Bacillota dominating, whereas Pseudomonadota prevailed in all control samples. Maroon 3 soils had higher diversity, but Cyanobacteria and Actinomycetota, dominant in controls, were replaced by Chloroflexota, Gemmatimonadota, and Acidobacteriota in polluted soils. At the genus level, Bacillus, Lysinibacillus, Virgibacillus, Brevibacillus, and Paenibacillus showed increased abundance in contaminated soils. Real-time PCR of alkB and C23DO genes indicated enhanced hydrocarbon degradation potential. FAPROTAX and PICRUSt2 analyses revealed enhanced microbial capacity for hydrocarbon degradation in polluted soils, with enriched functions related to chemoheterotrophy, aromatic compound degradation, and increased levels of alkane 1-monooxygenase, alcohol dehydrogenase, and protocatechuate 4,5-dioxygenase subunits. The findings highlight crude oil's impact on microbial community structure, reducing archaea and emphasizing bacterial dominance while underscoring shifts in microbial responses and functional gene expression in hydrocarbon degradation.
Keywords: C23DO; alkB; 16S rRNA metagenomics; Crude oil-contaminated soils; Real-time PCR.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
References
-
- Somee, M. R. et al. Halophiles in bioremediation of petroleum contaminants: challenges and prospects". In Bioremediation Environ Sustain (ed. Somee, M. R.) (Elsevier, 2021).
-
- Liu, X. et al. Microbial Remediation of Crude Oil in Saline Conditions by Oil-Degrading Bacterium Priestia megaterium FDU301. Appl. Biochem. Biotechnol.196(5), 2694–2712. 10.1007/s12010-022-04245-4 (2024). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

