Machine learning unveils hypoxia-immune gene hub for clinical stratification of thyroid-associated ophthalmopathy
- PMID: 40596187
- PMCID: PMC12214582
- DOI: 10.1038/s41598-025-05107-9
Machine learning unveils hypoxia-immune gene hub for clinical stratification of thyroid-associated ophthalmopathy
Abstract
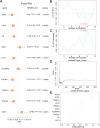
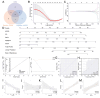
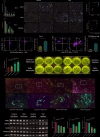
Thyroid-associated ophthalmopathy (TAO) is an autoimmune disorder affecting the orbit, potentially resulting in blindness. This study focused on the role of hypoxia in its pathogenesis through integrative bioinformatics and experimental validation. Five differentially expressed genes associated with hypoxia (HRDEGs) were identified via Gene Expression Omnibus (GEO) database mining: AGO2, CP, DIO3, PSMD14, WTIP. qPCR and immunohistochemistry confirmed reduced expressions of AGO2 and PSMD14, and elevated expression of DIO3 in TAO orbital tissues. Hypoxia exposure aggravated the above dysregulation and promoted proliferation and adipogenesis of orbital fibroblasts. A predictive model was developed using four machine learning algorithms and validated for its effectiveness in diagnosing TAO and assessing disease severity. Functional enrichment revealed hypoxia response, apoptosis, and programmed cell death. Protein-protein interaction and mRNA interaction networks of HRDEGs were established, predicting transcription factors, microRNAs, RNA-binding proteins, and drugs interacting with them. Immune infiltration analysis demonstrated the accumulation of Type 17 T helper cells and CD56 dim natural killer cells in high-risk patients, correlating with DIO3 upregulation and AGO2 downregulation. Flow cytometry confirmed the enrichment of these two cell types in the orbital tissue of TAO. This study revealed hypoxia-immunity crosstalk in TAO pathogenesis, providing a validated predictive model and molecular targets for precision interventions.
Keywords: Hypoxia; Immune microenvironment; Predictive model; Thyroid-associated ophthalmopathy.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

