Analysis of molecular and cellular bases of honey bee mushroom body development
- PMID: 40596271
- PMCID: PMC12215803
- DOI: 10.1038/s41598-025-06268-3
Analysis of molecular and cellular bases of honey bee mushroom body development
Abstract
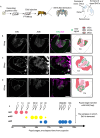
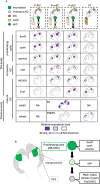
In the honey bee, mushroom bodies (MBs), a higher-order center of the insect brain, comprise three class I Kenyon cell (KC) subtypes (lKC, mKC, and sKC) with distinct somata sizes and locations and gene expression profiles. While these KC subtypes have been suggested to function in different behavioral regulations, the molecular and cellular basis of their development remains obscure. Here, we showed that lKCs, mKCs, and sKCs are produced in that order at different pupal stages by labeling proliferating MB cells with 5-ethynil-2'-deoxyuridine at various pupal stages. RNA-sequencing analysis of FACS-sorted pupal MB cells identified genes that were upregulated in proliferating and non-proliferating MB cells, respectively. Furthermore, in situ hybridization of some of these genes labeled the proliferating cells or immature KCs in the MBs at pupal stages producing each subtype. We found that the expression patterns of SoxNeuro, optix, and asense were consistent with those in Drosophila MBs, while odd-paired, which functions in neuroblasts in Drosophila, was preferentially expressed in immature KCs in honey bees. Our findings revealed the basic scheme of the molecular and cellular processes of honey bee MB development and suggested that they are at least partially different from those of Drosophila MB development.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
-
- Seeley, T. D. Honey bee ecology: A study of adaptation in social life (Princeton University Press, 2014).
-
- von Frisch, K. The Dance Language and Orientation of Bees (Harvard University Press, 1967).
-
- Johnson, B. R. Within-nest temporal polyethism in the honey bee. Behav. Ecol. Sociobiol.62, 777–784. 10.1007/s00265-007-0503-2 (2008).
-
- Heisenberg, M. Pattern recognition in insects. Curr. Opin. Neurobiol. 5, 475–481. 10.1016/0959-4388(95)80008-5 (1995). - PubMed
-
- Menzel, R. et al. The knowledge base of bee navigation. J. Exp. Biol.199, 141–146. 10.1242/jeb.199.1.141 (1996). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

