Regulatory T cells and matrix-producing cancer associated fibroblasts contribute on the immune resistance and progression of prognosis related tumor subtypes in ccRCC
- PMID: 40596367
- PMCID: PMC12214632
- DOI: 10.1038/s41598-025-06051-4
Regulatory T cells and matrix-producing cancer associated fibroblasts contribute on the immune resistance and progression of prognosis related tumor subtypes in ccRCC
Abstract
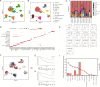
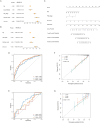
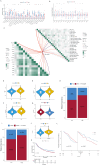
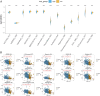
Clear cell renal cell carcinoma (ccRCC) is a prevalent malignant tumor in the field of urology. The effect of cell heterogeneity on the prognosis and reaction to treatment of ccRCC in large populations is still unclear. By analyzing public single cell RNA-sequencing and bulk RNA-sequencing data with the Scissor algorithm, we have identified three distinct prognosis related cancer cell subtypes which play an indispensable role on tumor metastasis, immune response and proliferation respectively. Besides, regulatory T cells (Tregs) and matrix producing cancer associated fibroblasts (matCAFs) were also recognized as crucial cell subtypes in the tumor microenvironment (TME). Moreover, potential interactions between Scissor + cells and other cells in TME were investigated to uncover regulatory mechanisms via 'Cell Chat' and cell2location algorithm. It is interesting that the interferon gamma signaling pathway and p53 signaling pathway contribute to the Scissor + transition of Tregs and matCAFs. The distinct activated transcription factor patterns were uncovered as well as the essential ligand-receptor pairs in the interactions among different cell subtypes, such as CXCL12-CXCR4 and COL6A2-SDC4. Then, we developed a risk score signature consisting of 10 genes, utilizing a 101-combination machine learning computational framework, which showed promising results in predicting the prognosis of patients. Furthermore, our study revealed variations in immune cell infiltration and the expression of immune related factors within the tumor microenvironment between different risk score groups, as well as the different sensitivity to the immunotherapy. In the end, we suggested Rapamycin as the additional therapy for the advanced ccRCC. In conclusion, our study created a signature to provide opportunities for predicting prognosis and improving treatments of ccRCC.
Keywords: Clear cell renal carcinoma; Machine learning; Multi-omics; Scissor.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin.71, 209–249. 10.3322/caac.21660 (2021). - PubMed
-
- Porta, C. et al. Pharmacotherapy for treating metastatic clear cell renal cell carcinoma. Expert Opin. Pharmacother.18, 205–216. 10.1080/14656566.2017.1282462 (2017). - PubMed
-
- Kotecha, R. R. et al. Towards individualized therapy for metastatic renal cell carcinoma. Nat. Rev. Clin. Oncol.16, 621–633. 10.1038/s41571-019-0209-1 (2019). - PubMed
-
- Gavish, A. et al. Hallmarks of transcriptional intratumour heterogeneity across a thousand tumours. Nature618, 598–606. 10.1038/s41586-023-06130-4 (2023). - PubMed
-
- Gonzalez-Silva, L. et al. Tumor Functional Heterogeneity Unraveled by scRNA-seq Technologies. Trends Cancer6, 13–19. 10.1016/j.trecan.2019.11.010 (2020). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

