Flavonoid biomarkers and co-expression gene networks characterized in ten Hemerocallis citrina accessions via multi-omics
- PMID: 40596419
- PMCID: PMC12216330
- DOI: 10.1038/s41598-025-06659-6
Flavonoid biomarkers and co-expression gene networks characterized in ten Hemerocallis citrina accessions via multi-omics
Abstract
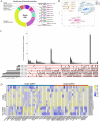
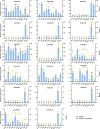
Hemerocallis citrina Baroni (H. citrina), a traditional medicinal and edible plant, is rich in bioactive flavonoids. This study investigated flavonoid metabolites in H. citrina flowers from ten accessions using UPLC-MS/MS-based metabolomics. A total of 642 flavonoids and 14 tannins were identified in the flowers. Among these, 375 flavonoids were classified as flavonols and flavone, representing for 39.33% and 17.53%, respectively. The identification of 309 differential flavonoid metabolites was achieved through analyses of nine pairwise comparison groups. These differential flavonoids in flowers of H. citrina could be distinctly categorized into two groups, clearly separating the 7 edible accessions from the 3 non-edible accessions. Significant variations in 14 flavonoid markers were observed among 10 H. citrina accessions originating from different regions, as determined by hierarchical clustering and ROC analyses. Transcriptomic analyses revealed that the majority of differentially expressed genes were enriched in the flavonoid metabolism pathway among these accessions. Integrated transcriptomic and metabolic analyses identified 14 differential flavonoid metabolites (DFMs) and 47 differentially expressed genes (DEGs) associated with flavonoid biosynthesis through Pearson's correlation analysis and WGCNA analyses. qRT-PCR validation of 13 DEGs confirmed the consistency of transcriptomic data. A flavonoid-gene correlation network indicated that the 14 DFMs might be directly regulated by 17 DEGs, comprising 13 flavnoid metabolism-related genes and 4 transcript factors. These findings provide biological and chemical insights into flavonoid metabolic difference across H. citrina origins, offering a theoretical basis for food and medical applications, and enabling clear differentiation between edible and non-edible H. citrina flowers in the market.
Keywords: Hemerocallis citrina; Correlation network; Differentially expressed genes; Flavonoid metabolism; Flowers.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical statement: This statement pertains to research, collection, or utilization activities involving the Hemerocallis citrina species. We strictly comply with the Wild Plant Protection Law of the People’s Republic of China, respecting and protecting wild plant resources while utilizing their natural value responsibly. The plant resources were identified by Dr. Tao Ma and are stored at Location Shanxi Agricultural University, Shanxi Academy of Agricultural Science, High Latitude Crops Institution (Geographical Coordinates: 113° 20′ 50.784" E, 40° 1′ 34.748" N). All materials used in this study, with the exception of MKLD and HXK (currently under variety registration application), have been officially registered in the National Crop Germplasm Resources Registration System of China ( http://111.203.21.74:8180/#/login?redirect=%2FresourcesInfo ) (registration numbers: DTHH: GHHC1240000006; SWJZ: GHHC1240000041; DWZ: GHHC1240000063; MZH: GHHC1240000018; CLH: GHHC1240000049; QXH: GHHC1240000140; MLH: GHHC1240000040; ZZN1: GHHC1240000103). However, these registered germplasm resources are strictly protected and not available for utilization or sharing.
Figures




References
-
- Liu, W. et al. Study the effects of drying processes on chemical compositions in daylily flowers using flow injection mass spectrometric fingerprinting method and chemometrics. Food Res. Int.102, 493–503 (2017). - PubMed
-
- Ma, T. et al. Qualitative and quantitative analysis of the components in flowers of Hemerocallis citrina Baroni by UHPLC–Q-TOF-MS/MS and UHPLC–QQQ-MS/MS and evaluation of their antioxidant activities. J. Food Compos. Anal.120, 105329 (2023).
-
- Wang, Y. et al. Advances in researches on chemical composition and functions of Hemerocallis plants. Medicinal plant9(2), 16–21 (2018).
-
- Ma, T. et al. Chemical constituents and mechanisms from Hemerocallis citrina Baroni with anti-neuroinflammatory activity. J. Funct. Foods102, 105427 (2023).
MeSH terms
Substances
Grants and funding
- sxzyk202409/Conservation and Utilization of Hemerocallis citrina Baroni Germplasm Resources in Shanxi Province
- 2024020/Research on New Techniques for Facility Cultivation of Datong Daylily Tissue-cultured Seedlings
- YZGC153/Germplasm Innovation and Breeding of Late-maturing Hemerocallis citrina Baroni Varieties
LinkOut - more resources
Full Text Sources

