Genetic association analysis between LDL-c lowering drugs and portal hypertension using Mendelian randomization analysis
- PMID: 40596422
- PMCID: PMC12217002
- DOI: 10.1038/s41598-025-08153-5
Genetic association analysis between LDL-c lowering drugs and portal hypertension using Mendelian randomization analysis
Abstract
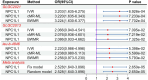
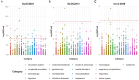
Clinical guidelines recommend the use of statins to reduce portal pressure and alleviate portal hypertension (PH). However, there is a lack of population-level studies on the use of non-statin Low-Density Lipoprotein Cholesterol (LDL-c) reduction agents for the treatment of PH. This study utilized a novel method, Mendelian Randomization (MR) analysis, to investigate the impact of commonly used LDL-c-lowering medications on PH. Instrumental variables (IVs) for eight lipid-lowering drug-related genes were extracted from three large-scale LDL-c databases of Genome-Wide Association Studies (GWAS), followed by MR analysis. The MR results indicated that, compared to normal individuals, lower expression of CETP and NPC1L1 in whole blood (result of meta-analysis: CETP [OR: 0.322, 95%CI:0.130-0.795, P = 1.396e-02], NPC1L1 [OR: 0.057, 95%CI: 0.022-0.146, P = 2.670e-09]) is associated with reduced portal pressure. The IVs of target genes were subjected to MR analysis with coronary atherosclerosis (CAD) as a positive control, confirming that the IVs can effectively substitute for the biological function of the target gene, thereby further enhancing the reliability of the results. Subsequently, Summary-based Mendelian Randomization (SMR) analysis was conducted by using expression quantitative trait loci (eQTL) data to validate the results of the MR analysis. The SMR results suggested that only NPC1L1 is associated with PH (OR: 0.648, 95%CI: 0.472-0.891, PSMR = 7.502e-3, PHEIDI = 0.747) from genetic correlation. Additionally, mediation analysis indicates that the therapeutic effect of NPC1L1 inhibitors on PH is partially mediated by tissue factor (mediating effect accounted for 18.52%, and the P value was 0.01). Phenome-Wide MR indicated that NPC1L1 inhibitors may be associated with 23 diseases or symptoms. In addition, NPC1L1 had genetic correlation between and alkaline phosphatase as well as total bilirubin, but no genetic correlation with alanine aminotransferase, aspartate aminotransferase, direct bilirubin, or gamma glutamyltransferase. In conclusion, this study systematically analyzed the genetic correlation between lipid-lowering drug targets and PH. From a genetic correlation perspective, we revealed that the potential therapeutic effect of NPC1L1 on PH may not be mediated through the reduction of LDL-c but rather through the modulation of tissue factors. Additionally, the potential side effects associated with NPC1L1 inhibition also were explored.
Keywords: Anticoagulation; Drug-target Mendelian randomization; Lipid-lowering drugs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
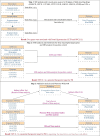
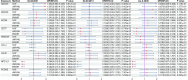
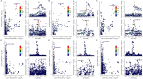
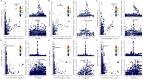
Figures





Similar articles
-
[Multi-omics Mendelian randomization study on the causality between non-ionizing radiation and facial aging].Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2025 Jun 20;41(6):594-603. doi: 10.3760/cma.j.cn501225-20240830-00320. Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2025. PMID: 40588408 Free PMC article. Chinese.
-
The causal association between lipid-lowering strategies and risk of intracranial aneurysms: A drug-target Mendelian randomization study.J Clin Lipidol. 2025 May-Jun;19(3):670-678. doi: 10.1016/j.jacl.2025.01.003. Epub 2025 Jan 22. J Clin Lipidol. 2025. PMID: 39979134
-
Eliciting adverse effects data from participants in clinical trials.Cochrane Database Syst Rev. 2018 Jan 16;1(1):MR000039. doi: 10.1002/14651858.MR000039.pub2. Cochrane Database Syst Rev. 2018. PMID: 29372930 Free PMC article.
-
Associations between lipid-lowering drugs and pregnancy and perinatal outcomes: a Mendelian randomization study.J Hypertens. 2024 Apr 1;42(4):727-734. doi: 10.1097/HJH.0000000000003664. Epub 2024 Jan 15. J Hypertens. 2024. PMID: 38230624
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

