The long-term effects of chemotherapy on normal blood cells
- PMID: 40596443
- PMCID: PMC12283364
- DOI: 10.1038/s41588-025-02234-x
The long-term effects of chemotherapy on normal blood cells
Erratum in
-
Author Correction: The long-term effects of chemotherapy on normal blood cells.Nat Genet. 2025 Aug;57(8):2075. doi: 10.1038/s41588-025-02315-x. Nat Genet. 2025. PMID: 40745026 Free PMC article. No abstract available.
Abstract
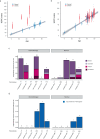
Several chemotherapeutic agents act by increasing DNA damage in cancer cells, triggering cell death. However, there is limited understanding of the extent and long-term consequences of collateral DNA damage in normal tissues. To investigate the impact of chemotherapy on mutation burdens and the cell population structure of normal tissue, we sequenced blood cell genomes from 23 individuals aged 3-80 years who were treated with a range of chemotherapy regimens. Substantial additional somatic mutation loads with characteristic mutational signatures were imposed by some chemotherapeutic agents, but the effects were dependent on the drug and blood cell types. Chemotherapy induced premature changes in the cell population structure of normal blood, similar to those caused by normal aging. The results show the long-term biological consequences of cytotoxic agents to which a substantial fraction of the population is exposed as part of disease management, raising mechanistic questions and highlighting opportunities for the mitigation of adverse effects.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: G.J.D. and U.M. are employees of and shareholders in AstraZeneca. M.R.S. and P.J.C. are cofounders of and shareholders in Quotient Therapeutics. The other authors declare no competing interests.
Figures













References
-
- DeVita, V. T. Jr. & Chu, E. A history of cancer chemotherapy. Cancer Res.68, 8643–8653 (2008). - PubMed
-
- Kollmannsberger, C., Hartmann, J. T., Kanz, L. & Bokemeyer, C. Risk of secondary myeloid leukemia and myelodysplastic syndrome following standard-dose chemotherapy or high-dose chemotherapy with stem cell support in patients with potentially curable malignancies. J. Cancer Res. Clin. Oncol.124, 207–214 (1998). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

