Leveraging chorionic villus biopsies for the derivation of patient-specific trophoblast stem cells
- PMID: 40596474
- PMCID: PMC12215853
- DOI: 10.1038/s42003-025-08393-1
Leveraging chorionic villus biopsies for the derivation of patient-specific trophoblast stem cells
Abstract
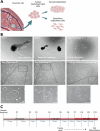
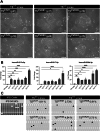
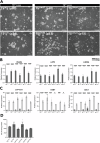
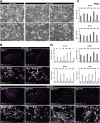
Human trophoblast stem (TS) cells are informative in vitro models for the generation and testing of biologically meaningful hypotheses. The goal of this project was to derive patient-specific TS cell lines from clinically available chorionic villus biopsies. Cell outgrowths were captured from human chorionic villus tissue specimens cultured in modified human TS cell medium. Cell colonies emerged early during the culture and cell lines were established and passaged for several generations. Karyotypes of the newly established chorionic villus-derived trophoblast stem (TSCV) cell lines were determined and compared to initial genetic diagnoses from freshly isolated chorionic villi. Phenotypes of TSCV cells in the stem state and following differentiation were compared to cytotrophoblast-derived TS (TSCT) cells. TSCV and TSCT cells uniformly exhibited similarities in the stem state and following differentiation into syncytiotrophoblast and extravillous trophoblast cells. Chorionic villus tissue specimens provide a valuable source for TS cell derivation. They expand the genetic diversity of available TS cells and are associated with defined clinical outcomes. TSCV cell lines provide a new set of experimental tools for investigating trophoblast cell lineage development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Update of
-
Leveraging chorionic villus biopsies for the derivation of patient-specific trophoblast stem cells.medRxiv [Preprint]. 2024 Jul 23:2022.12.07.22283218. doi: 10.1101/2022.12.07.22283218. medRxiv. 2024. Update in: Commun Biol. 2025 Jul 1;8(1):964. doi: 10.1038/s42003-025-08393-1. PMID: 39108523 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- R01 HD112559/HD/NICHD NIH HHS/United States
- R01 HD020676/HD/NICHD NIH HHS/United States
- R01 HD099638/HD/NICHD NIH HHS/United States
- F32HD096809/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01 AI154535/AI/NIAID NIH HHS/United States
- K99 HD115834/HD/NICHD NIH HHS/United States
- K99HD107262/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD020676/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD099638/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD105734/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- AI154535/U.S. Department of Health & Human Services | NIH | Office of Research on Women's Health (NIH Office of Research on Women's Health)
- F32 HD096809/HD/NICHD NIH HHS/United States
- R01 HD105734/HD/NICHD NIH HHS/United States
- K99 HD107262/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

