Gut microbiota constituents may affect hypertrophic scarring risk through interaction with specific immune cells in a two-step, two-sample Mendelian randomization study
- PMID: 40596527
- PMCID: PMC12216485
- DOI: 10.1038/s41598-025-07455-y
Gut microbiota constituents may affect hypertrophic scarring risk through interaction with specific immune cells in a two-step, two-sample Mendelian randomization study
Abstract
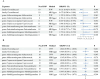
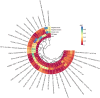
Hypertrophic scars (HS), classified as abnormal scarring, result from an overactive tissue response during wound healing following dermal trauma. Nonetheless, the precise mechanistic pathway underlying its occurrence remains elusive. The principal aim of this study is to elucidate the causal relationship among gut microbiota (GM), immune cell function, and hypertrophic scarring in a European demographic. Leveraging the genome-wide association analysis (GWAS) database, we conducted a two-sample Mendelian randomization (MR) study on gut microbiota (GM), immune cells, and HS. To ascertain the causality between GM, immune cells, and HS, we utilized the inverse variance weighted (IVW) method while employing multiple approaches to negate the effects of pleiotropy and heterogeneity. Furthermore, we quantitatively evaluated the influences of immune cells-mediated GM on hypertrophic scar through a two-step MR analysis. The two-sample MR analysis demonstrated a potential causality between 5 genera of gut microbiotas and 23 immune cell traits with respect to hypertrophic scarring. Further, our results showed that the causal pathway from the genus Subdoligranulum to hypertrophic scar (HS) was mediated by B cell-activating factor receptor (BAFF-R) on CD20- CD38- B cell, with a beta value of 0.034 (95% CI [0.002, 0.066]; P = 0.004), contributing to 7.60% of the total effect of Subdoligranulum on HS. Similarly, CD24 on IgD + CD38 + B cell exhibited a causal impact in the pathway from genus Coprococcus 1 to HS, with a beta value of -0.015 (95% CI [-0.029, -0.001]; P = 0.023), constituting 6.70% of the total effect of Coprococcus 1 on HS. Additionally, the CD8 + T cell %T cell mediated the causal pathway from the genus Adlercreutzia to HS with a beta value of 0.075 (95% CI [0.017, 0.133]; P = 0.024), contributing to 10.10% of the total effect of Adlercreutzia on HS. Our study indicates that the development of hypertrophic scars might be influenced by specific gut microbiota and immune cells. We highlight the possible role of two distinct immune cell genotypes as mediators in this relationship. However, most statistical significance of these findings was not maintained after FDR correction, suggesting our results should be viewed as preliminary and interpreted with caution. Further research is needed to confirm these associations.
Keywords: Gut microbiota; Hypertrophic scar; Immune cells; Mendelian randomization; Two-step analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Each study incorporated in the GWAS used in the present study was approved by local research ethics committees or Institutional Review Boards, and all participants had given their informed consent. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
CD27 on IgD-CD38-B Cells Mediates the Coprococcus-COPD Link.Int J Chron Obstruct Pulmon Dis. 2025 Jul 3;20:2173-2182. doi: 10.2147/COPD.S518455. eCollection 2025. Int J Chron Obstruct Pulmon Dis. 2025. PMID: 40626314 Free PMC article.
-
Inflammatory cytokines mediate the gut microbiota-EGPA subtype link: a Mendelian randomization study.Clin Rheumatol. 2025 Jul;44(7):3061-3071. doi: 10.1007/s10067-025-07526-5. Epub 2025 Jun 12. Clin Rheumatol. 2025. PMID: 40500572
-
Increased risk of hypertrophic scarring in estrogen receptor-positive breast cancer: a bidirectional mendelian randomization study.Arch Dermatol Res. 2025 Jan 20;317(1):299. doi: 10.1007/s00403-024-03737-z. Arch Dermatol Res. 2025. PMID: 39833481
-
Silicone gel sheeting for treating hypertrophic scars.Cochrane Database Syst Rev. 2021 Sep 26;9(9):CD013357. doi: 10.1002/14651858.CD013357.pub2. Cochrane Database Syst Rev. 2021. PMID: 34564840 Free PMC article.
-
Interventions for acne scars.Cochrane Database Syst Rev. 2016 Apr 3;4(4):CD011946. doi: 10.1002/14651858.CD011946.pub2. Cochrane Database Syst Rev. 2016. PMID: 27038134 Free PMC article.
References
-
- Trujillo Cubillo, L., Gurdal, M. & Zeugolis, D. I. Corneal fibrosis: from in vitro models to current and upcoming drug and gene medicines. Adv. Drug Deliv. Rev.209, 115317. 10.1016/j.addr.2024.115317 (2024). - PubMed
MeSH terms
Grants and funding
- 2023HMZD07/HwaMei Reasearch Foundation of Ningbo No.2 Hospital
- 2023HMZD07/HwaMei Reasearch Foundation of Ningbo No.2 Hospital
- 2023RC081/Medical Scientific Reasearch Foundation of Zhejiang Province
- 2022498346/Medical Scientific Reasearch Foundation of Zhejiang Province
- 2023HMJQ25/Zhu Xiu Shan Talent Project of Ningbo No.2 Hospital
LinkOut - more resources
Full Text Sources
Research Materials

