QTL mapping for yield contributing traits in mungbean (Vigna radiata L.) using a RIL population
- PMID: 40596720
- PMCID: PMC12218555
- DOI: 10.1038/s41598-025-99687-1
QTL mapping for yield contributing traits in mungbean (Vigna radiata L.) using a RIL population
Abstract
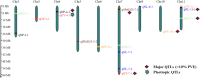
Mungbean (Vigna radiata L.) is one of the most important yet genomically under-researched leguminous food crop. Its productivity is low due to the complex nature of yield realization, which is regulated by various yield-contributing traits. Thus, understanding the genetic basis of these traits is essential for developing an ideal genotype with high yield. In this study, mapping of quantitative trait loci (QTLs) was conducted for six yield-contributing traits using a recombinant inbred line (RIL) population (166 number), developed by crossing two contrasting genotypes (Pusa Baisakhi × PMR-1). The genotyping-by-sequencing (GBS) of RILs was used to construct the genetic map using 1347 single nucleotide polymorphism (SNPs). The QTL mapping using multiple interval mapping (MIM) and composite interval mapping (CIM) has identified 17 yield contributing QTLs of which, 4 for number of leaves/plant (NL), 3 for plant height (PH), 2 for SPAD value, 3 for 100 seed weight (SW), 2 for number of pods/plant (NP), and 3 for total grain yield (GY). The Logarithm of Odds (LOD) scores for these QTLs ranged from ~ 3-9, while phenotypic variance explained (PVE) ranged from ~ 9-24%. Several candidate genes with mRNA expression and protein-altering mutations were identified as having a direct role in key processes like growth (LOC106756212, LOC106776425, LOC106777991), flowering (LOC106777903, LOC106768860), metabolism (LOC106757749, LOC106758189), etc. The candidate genes are validated through digital gene expression analysis. In addition, Insertion-Deletion (InDel) markers were also developed for the identified QTLs which hold broad applications for the improvement of yield-related traits in mungbean.
Keywords: Candidate genes; Composite interval mapping; Genotyping by sequencing; Multiple interval mapping; Yield traits.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Comprehensive genetic dissection of yield-related traits utilizing quantitative trait loci sequencing approach in mungbean.J Plant Physiol. 2025 Sep;312:154552. doi: 10.1016/j.jplph.2025.154552. Epub 2025 Jun 27. J Plant Physiol. 2025. PMID: 40614315
-
Quantitative trait locus mapping for salt and drought tolerance traits in wheat (Triticum aestivum L.).BMC Plant Biol. 2025 Jul 1;25(1):787. doi: 10.1186/s12870-025-06774-6. BMC Plant Biol. 2025. PMID: 40596820 Free PMC article.
-
Identification of quantitative trait loci and candidate genes underlying kernel traits of wheat (Triticum aestivum L.) in response to drought stress.Theor Appl Genet. 2025 Aug 19;138(9):216. doi: 10.1007/s00122-025-05001-y. Theor Appl Genet. 2025. PMID: 40828177
-
Selegiline for Alzheimer's disease.Cochrane Database Syst Rev. 2003;(1):CD000442. doi: 10.1002/14651858.CD000442. Cochrane Database Syst Rev. 2003. PMID: 12535396
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
-
- Dikshit, H.K., Mishra, G.P., Somta, P., Shwe, T., Alam, A.K.M.M. & Nair, R.M. Classical genetics and traditional breeding in mungbean. In (Nair, R., Schafleitner, R., Lee, S.H. eds.) The Mungbean Genome. Compendium of Plant Genomes. 43–54. 10.1007/978-3-030-20008-4_4. ISBN: 978-303020008-4 (Springer, 2020).
-
- Mishra, G.P., Priti, Dikshit, H.K., Aski, M., Sangwan, S., Stobdan, T. & Praveen, S. Microgreens: A novel food for nutritional security. In Conceptualizing Plant-Based Nutrition: Bioresources, Nutrients Repertoire and Bioavailability. 123–156. (Springer, 2022).
-
- Priti, Mishra, G.P., Dikshit, H.K., Vinutha, T., Mechiya, T., Stobdan, T. & Praveen, S. Diversity in phytochemical composition, antioxidant capacities, and nutrient contents among mungbean and lentil microgreens when grown at plain-altitude region (Delhi)and high-altitude region (Leh-Ladakh), India. Front. Plant Sci.12, 710812 (2021). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

