Core histone families of mollusca: systematic identification, evolutionary insights, and functional analysis
- PMID: 40596809
- PMCID: PMC12210499
- DOI: 10.1186/s12864-025-11776-7
Core histone families of mollusca: systematic identification, evolutionary insights, and functional analysis
Abstract
Background: Histones are the basic packaging units of eukaryotic DNA and are essential for the dynamics of chromatin and the regulation of epigenetics. Canonical histones and their variants exhibit important functional differences in biological processes. However, little is known about the role of histone family members in molluscs, which are known for their ecological and morphological diversity.
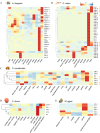
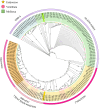
Results: Core histone families of 28 molluscan species (12 bivalves, 8 gastropods, 6 cephalopods, 1 scaphopod and 1 polyplacophora) were systematically identified. The evolutionary conservation and lineage-specific innovations were discovered using phylogenomic and transcriptomic analyses. Cephalopods showed a striking expansion of canonical histone genes with brain-enriched expression patterns. Synteny analyses revealed conserved, collinear histone clusters unique to cephalopods. Histone variants, specially H2A and H3 paralogs, display conserved motifs potentially involved in nucleosome stability and lineage-specific residues involved in functional specialization. Developmental transcriptomics revealed the dynamic expression of histone variants in early embryogenesis and the gonads, suggesting that H2A and H3 variants are involved in chromatin remodeling, pluripotency maintenance and germline regulation. Macro-H2A was highly expressed during larval neurodevelopment and in sensory organs, suggesting important roles in neural plasticity.
Conclusion: This study represents the first comprehensive inventory and characterization of core histone genes in molluscs, and will facilitate understanding of the evolutionary patterns and functional properties of core histones in relation to neurogenesis of molluscs. These findings advance our understanding of chromatin evolution and its contribution to phenotypic innovation in non-model taxa.
Keywords: Core histone families; Expression; Mollusca; Nervous system; Systematic characterization.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All our experiments have complied with relevant institutional, national, and international guidelines and legislation. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. - PubMed
-
- Millán-Zambrano G, Burton A, Bannister AJ, Schneider R. Histone post-translational modifications - cause and consequence of genome function. Nat Rev Genet. 2022;23(9):563–80. - PubMed
-
- Waterborg JH. Evolution of histone H3: emergence of variants and conservation of post-translational modification sites. Biochem Cell Biol. 2012;90(1):79–95. - PubMed
MeSH terms
Substances
Grants and funding
- SDBX202302013/Shandong Postdoctoral Science Foundation
- 2022KJ046/Youth Innovation Team Plan of Higher Education Institutions in Shandong Province
- 32422088/National Natural Science Foundation of China
- No. 202341009/Fundamental Research Funds for the Central Universities
- CARS-49/Modern Agro-industry Technology Research System
LinkOut - more resources
Full Text Sources

