Nanopore-based targeted sequencing (NTS) for drug-resistant tuberculosis: an integrated tool for personalized treatment strategies and guidance for new drug development
- PMID: 40596923
- PMCID: PMC12220424
- DOI: 10.1186/s12879-025-11227-4
Nanopore-based targeted sequencing (NTS) for drug-resistant tuberculosis: an integrated tool for personalized treatment strategies and guidance for new drug development
Abstract
Background: Drug-resistant tuberculosis has emerged as a major public health issue that requires immediate attention. NTS is an innovative method that allows for the direct detection of clinical samples without the need for culture. It could provide more accurate, reliable, and comprehensive information on drug resistance.
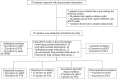
Methods: We collected clinical data retrospectively from patients suspected of having drug-resistant tuberculosis who visited the tuberculosis department at the Second Hospital of Nanjing in Jiangsu Province, China, from December 2023 to December 2024. The diagnostic efficiency of NTS for different types of drug-resistant tuberculosis and antimicrobial resistance was calculated. The relationship between resistance genes, mutated amino acids, and mutation sites was demonstrated.
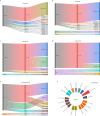
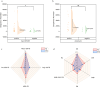
Results: In this study, a total of 107 patients with drug-resistant tuberculosis were included, comprising 43 cases of mono-drug resistant tuberculosis, 20 patients with poly-drug resistant tuberculosis, 22 cases of multidrug-resistant tuberculosis, 21 cases of pre-extensively drug-resistant tuberculosis and 1 case of extensively drug-resistant tuberculosis. The accuracy of NTS in diagnosing drug-resistant tuberculosis ranged from 42.9 to 93.0%. Except for second-line injectable drugs, NTS achieved a sensitivity of over 70% for other anti-tuberculosis drugs. Serine was identified as the most frequently mutated amino acid in both the rpoB gene (66.2%, 49/74) and the katG gene (86.3%, 44/51). Additionally, the most frequently mutated amino acids in the embB gene, rpsL gene, and gyrA gene were methionine (94.7%, 44/51), lysine (100%, 28/28), and aspartic acid (66.7%, 20/30), respectively.
Conclusion: NTS could effectively and precisely deliver comprehensive drug resistance results, assisting medical professionals to create more personalized treatment plans. Besides, it would encourage the development of new anti-tuberculosis drugs to broaden clinical treatment options for drug-resistant tuberculosis.
Keywords: Amino acids; Drug-resistant genes; Drug-resistant tuberculosis(DR-TB); Nanopore-based targeted sequencing(NTS); Targeted next-generation sequencing(tNGS).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study received approval from the Human Research Ethics and System Review Committee of the Second Hospital of Nanjing (ID: 2024-LS-ky026). The study protocol was consistent with the ethical guidelines of the 1975 Declaration of Helsinki. Consent to participate: Informed consent was obtained from all the participants. Consent for publication: Not applicable. Conflict of interest: All authors have no conflict of interest to declare. Clinical trial number: Not Applicable.
Figures

Similar articles
-
Prevalence and molecular characterization of drug-resistant Mycobacterium tuberculosis in Heyuan City in China.Front Cell Infect Microbiol. 2025 Jun 12;15:1586938. doi: 10.3389/fcimb.2025.1586938. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40575484 Free PMC article.
-
Targeted next-generation sequencing: a promising approach for Mycobacterium tuberculosis detection and drug resistance when applied in paucibacillary clinical samples.Microbiol Spectr. 2025 Jul;13(7):e0312724. doi: 10.1128/spectrum.03127-24. Epub 2025 Jun 10. Microbiol Spectr. 2025. PMID: 40492765 Free PMC article.
-
Evaluating culture-free targeted next-generation sequencing for diagnosing drug-resistant tuberculosis: a multicentre clinical study of two end-to-end commercial workflows.Lancet Infect Dis. 2025 Mar;25(3):325-334. doi: 10.1016/S1473-3099(24)00586-3. Epub 2024 Oct 29. Lancet Infect Dis. 2025. PMID: 39486428
-
Xpert® MTB/RIF assay for extrapulmonary tuberculosis and rifampicin resistance.Cochrane Database Syst Rev. 2018 Aug 27;8(8):CD012768. doi: 10.1002/14651858.CD012768.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2021 Jan 15;1:CD012768. doi: 10.1002/14651858.CD012768.pub3. PMID: 30148542 Free PMC article. Updated.
-
Xpert MTB/XDR for detection of pulmonary tuberculosis and resistance to isoniazid, fluoroquinolones, ethionamide, and amikacin.Cochrane Database Syst Rev. 2022 May 18;5(5):CD014841. doi: 10.1002/14651858.CD014841.pub2. Cochrane Database Syst Rev. 2022. PMID: 35583175 Free PMC article.
References
-
- Farhat M, Cox H, Ghanem M, Denkinger CM, Rodrigues C, Abd El Aziz MS, Enkh-Amgalan H, Vambe D, Ugarte-Gil C, Furin J, et al. Drug-resistant tuberculosis: a persistent global health concern. Nat Rev Microbiol. 2024;22(10):617–35. - PubMed
-
- Global. tuberculosis report 2024 [https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa...]
-
- Sanchini A, Lanni A, Giannoni F, Mustazzolu A. Exploring diagnostic methods for drug-resistant tuberculosis: A comprehensive overview. Tuberculosis (Edinb). 2024;148:102522. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

