Senescence-associated alterations in histone H3 modifications, HP1 alpha levels and distribution, and in the transcriptome of vascular smooth muscle cells in different types of senescence
- PMID: 40597225
- PMCID: PMC12220758
- DOI: 10.1186/s12964-025-02315-8
Senescence-associated alterations in histone H3 modifications, HP1 alpha levels and distribution, and in the transcriptome of vascular smooth muscle cells in different types of senescence
Abstract
Background: Cellular senescence is a fundamental process leading to organismal aging and age-related diseases. Alterations accompanying cellular senescence concern, among others, nucleus architecture, chromatin structure, DNA damage and gene expression. Some changes are universal for all types of senescence, but some characteristics are typical for a given senescence inductor or cell type. The aim of the study was to analyze senescence-associated alterations in chromatin modifications and look for differences depending on senescence type (replicative, RS and stress-induced premature senescence, SIPS) in vascular smooth muscle cells (VSMCs) in vitro. The alterations were compared with those observed in VSMCs derived from atherosclerotic plaques (ex vivo) and, to assess their universality, with those in senescent fibroblasts.
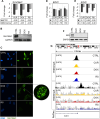
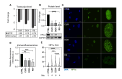
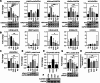
Methods: We investigated the level and distribution of HP1α and H3 modifications that are markers of hetero- and euchromatin (H3K9me3, H3K27me3, H3K4me3, H3K9Ac - WB and IF), alterations in the transcriptomic profile (DNA microarray, qPCR), H3K4me3, H3K9me3 and HP1α protein distribution in the genome (ChIP-seq), and expression of enzymes involved in histone post-translational modifications (DNA microarray, qPCR, WB, IF).
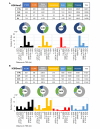
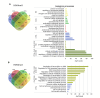
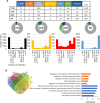
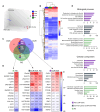
Results: Our results have shown that the decline in H3K4me3 and H3K9me3 modifications and in HP1α is a universal hallmark of senescence in all tested cell and senescence types, although the extent of the change depends on the senescence inductor. The distribution of H3K4me3 and H3K9me3 in the genome of VSMCs depends on the senescence type, and the transcriptomic analysis identified genes and processes specific to each type.
Conclusions: We characterized senescence and cell type-dependent changes in chromatin-associated proteins and enzymes involved in histone H3 decoration which, in consequence, impact senescence-associated gene expression. We can conclude that certain similar alterations occur in senescent VSMCs ex vivo, although inter-individual differences usually obscure them. Our results clearly showed that differences existed not only between young and senescent cells but also between SIPS and RS ones. The subtle differences between various SIPS types suggest that various stressors activate the same cellular mechanisms. This study can serve as a starting point to search for factors that may be used to distinguish between SIPS and RS, which in turn could be helpful in defining conditions responsible for accelerated aging.
Keywords: Chromatin structure; HP1α; Histone H3 modifications; Senescence; VSMCs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Research involving human biological material - smooth muscle cells isolated from atherosclerotic plaque were obtained from patients who underwent carotid endarterectomies. Studies with human samples were approved by the Ethical Committee of the Central Clinical Hospital Ministry of Internal Affairs (2017/114). Written informed consent was obtained from all patients. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023;186(2):243–78. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

