Clinical applications of and molecular insights from RNA sequencing in a rare disease cohort
- PMID: 40597352
- PMCID: PMC12210447
- DOI: 10.1186/s13073-025-01494-w
Clinical applications of and molecular insights from RNA sequencing in a rare disease cohort
Abstract
Background: RNA sequencing (RNA-seq) is emerging as a valuable tool for identifying disease-causing RNA transcript aberrations that cannot be identified by DNA-based testing alone. Previous studies demonstrated some success in utilizing RNA-seq as a first-line test for rare inborn genetic conditions. However, DNA-based testing (increasingly, whole genome sequencing) remains the standard initial testing approach in clinical practice. The indications for RNA-seq after a patient has undergone DNA-based sequencing remain poorly defined, which hinders broad implementation and funding/reimbursement.
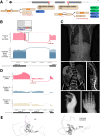
Methods: In this study, we identified four specific and familiar clinical scenarios, and investigated in each the diagnostic utility of RNA-seq on clinically accessible tissues: (i) clarifying the impact of putative intronic or exonic splice variants (outside of the canonical splice sites), (ii) evaluating canonical splice site variants in patients with atypical phenotypes, (iii) defining the impact of an intragenic copy number variation on gene expression, and (iv) assessing variants within regulatory elements and genic untranslated regions.
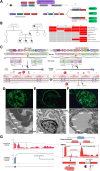
Results: These hypothesis-driven RNA-seq analyses confirmed a molecular diagnosis and pathomechanism for 45% of participants with a candidate variant, provided supportive evidence for a DNA finding for another 21%, and allowed us to exclude a candidate DNA variant for an additional 24%. We generated evidence that supports two novel Mendelian gene-disease associations (caused by variants in PPP1R2 and MED14) and several new disease mechanisms, including the following: (1) a splice isoform switch due to a non-coding variant in NFU1, (2) complete allele skew from a transcriptional start site variant in IDUA, and (3) evidence of a germline gene fusion of MAMLD1-BEND2. In contrast, RNA-seq in individuals with suspected rare inborn genetic conditions and negative whole genome sequencing yielded only a single new potential diagnostic finding.
Conclusions: In summary, RNA-seq had high diagnostic utility as an ancillary test across specific real-world clinical scenarios. The findings also underscore the ability of RNA-seq to reveal novel disease mechanisms relevant to diagnostics and treatment.
Keywords: Genetic disease; Molecular diagnostic techniques; Novel disease mechanisms; Pediatric rare disease; Putative (new) disease gene; RNA sequencing (RNA-seq); Splice variants; Transcriptomics; Variant of uncertain significance.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Participants were provided written informed consent under various research studies approved by the Research Ethics Board (REB) at The Hospital for Sick Children (SickKids), Toronto, Ontario, Canada. Written consent was obtained from each proband’s parents or guardians, with assent provided by the proband where appropriate. This consent included publication of de-identified clinical and research findings. The study was conducted in accordance with the Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans (TCPS 2), Ontario’s Personal Health Information Protection Act (PHIPA), SickKids institutional research ethics guidelines and conformed to the principles of the Declaration of Helsinki. Competing interests: The authors declare no competing interests.
Figures




References
-
- Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, et al. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare Mendelian disease. The American J Human Genet. 2019 Mar;104(3):466–83. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0002929719300126 - PMC - PubMed
-
- Kernohan KD, Boycott KM. The expanding diagnostic toolbox for rare genetic diseases. Nat Rev Genet. 2024;25(6):401-15. 10.1038/s41576-023-00683-w. - PubMed
-
- Jaramillo Oquendo C, Wai HA, Rich WI, Bunyan DJ, Thomas NS, Hunt D, et al. Identification of diagnostic candidates in Mendelian disorders using an RNA sequencing-centric approach. Genome Med. 2024 Sep;16(1):110. Available from: https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-024-013... - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

