Unraveling key transposable elements in pathogen-induced bovine mastitis through comparative in vivo and in vitro transcriptomic analysis
- PMID: 40597583
- PMCID: PMC12210757
- DOI: 10.1186/s12864-025-11740-5
Unraveling key transposable elements in pathogen-induced bovine mastitis through comparative in vivo and in vitro transcriptomic analysis
Abstract
Background: Bovine mastitis poses significant hazards to the yield and quality of dairy products, severely hindering the development of the dairy industry. Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) are well-established as two of the primary pathogens causing the disease. Transposable elements (TEs), occupying a notable proportion of livestock genomes, primarily function as regulatory elements modulating gene expression. Extensive studies have indicated that TEs contribute to transcriptional changes in the host during pathogen invasion. However, despite their potential significance, the key functional TEs associated with bovine mastitis remain unclear, highlighting the need to explore the critical roles of TEs in the immune processes of this disease.
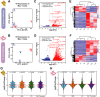
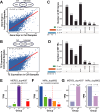
Results: In this study, in vitro and in vivo mastitis models were established using bovine mammary alveolar cells (Mac-T cells) and Chinese Holstein cows, respectively. In vitro findings showed distinct expression profiles of genes and TEs in response to challenges posed by S. aureus and E. coli. Specifically, 1,750 differentially expressed genes (DE Genes) and 3,046 differentially expressed TEs (DE TEs) were identified in the S. aureus challenge, while 2,353 DE Genes and 22,259 DE TEs were identified in the E. coli challenge. TEs were found to regulate the expression of genes primarily within immune-related pathways, including IL-17 and HIF-1 signaling pathways. TE-gene-QTL regulatory networks were established, providing preliminary insights into the molecular genetic mechanisms of TE regulation. By integrating in vitro and in vivo data, we identified and further validated two TE instances from MER53/DNA transposon and MIRc/SINE families as stably activated and repressed transcriptional markers for S. aureus mastitis, respectively.
Conclusions: Our research underscores the potential regulatory roles of TEs in the pathogenesis of bovine mastitis and highlights their applicability as molecular markers for early diagnosis and prevention of this economically significant disease. Our study offers novel insights for the breeding and improvement of resistance to pathogen-induced mastitis in dairy cattle.
Keywords: Escherichia coli; Staphylococcus aureus; Bovine mastitis; Transcriptomics; Transposable elements.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Animal data involved in this study originated from a previous study conducted in accordance with the local animal welfare guidelines. Since no additional animals were used, this study did not require a statement on ethics approval. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

