Analysis of genomic selection characteristics of local cattle breeds in Gansu
- PMID: 40597588
- PMCID: PMC12211739
- DOI: 10.1186/s12864-025-11753-0
Analysis of genomic selection characteristics of local cattle breeds in Gansu
Abstract
Background: The distinctive geography and climate of Gansu Province have given rise to three indigenous cattle breeds-Zaosheng, Anxi, and Yangba. Renowned for their superior meat quality and remarkable adaptability, these breeds are crucial for maintaining genetic diversity. However, they are under threat from intensive farming practices, environmental degradation, and genetic drift, which could lead to an irreversible loss of genetic resources. Thanks to natural and artificial selection, these breeds possess genetic markers that enhance their adaptation to extreme environments and improve key economic traits. By integrating comprehensive genome data from multiple breeds, this study aims to analyze population genetics, detect composite selection signals, and perform functional enrichment to uncover the mechanisms behind genetic differentiation and adaptive evolution. This research is pivotal for developing resilient breeds and ensuring sustainable resource management.
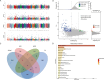
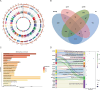
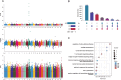
Results: The genetic background of local cattle breeds in Gansu shows a mix between indicine cattle (Bos indicus) and taurine cattle (Bos taurus), with geographical differentiation: Yangba cattle in the southeast mainly exhibit indicine ancestry (54.43%), while Anxi and Zaosheng cattle in the northwest show a predominance of taurine ancestry (86.51% and 74.81%, respectively). This divergence is closely related to historical ethnic migrations, geographic barriers, and gene flow along the Silk Road. Selection signal analysis has revealed specific adaptation mechanisms in different populations: Yangba cattle exhibit strong selection signals in the T-cell receptor pathway (FYN, FYB1) and skeletal development genes (SOX6), which may be related to their adaptation to hot and humid environments and mountainous terrain; Anxi cattle show adaptive evolution in nitrogen metabolism (CA8, CA10) and adherens junction pathways (CTNNA2), possibly reflecting the genetic basis for their adaptation to arid conditions; Zaosheng cattle display strong selection signals in muscle development (LARGE1, SGCZ) and immune regulation genes (SLAMF family), likely associated with enhanced meat production performance and increased pathogen resistance driven by artificial breeding.
Conclusion: This study explores the drivers of genetic diversity and adaptive evolution in Gansu's native cattle breeds, emphasizing the impact of geography and human activity on genetic divergence. It provides a theoretical basis for conserving breed resources, identifying functional genes, and developing breeding strategies.
Keywords: Gansu local breeds; Genetic evolution; Population genetics; Whole-genome re-sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All experimental procedures involving cattle were conducted in strict compliance with the Regulations for the Administration of Experimental Animals (approved by the State Council of the People’s Republic of China). Written informed consent was obtained from all cattle owners prior to sample collection. This study was formally approved by the Animal Administration and Ethics Committee of Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences (CAAS) (Permit No.: SYXK-2024-0023). Additionally, the study adheres to the guidelines outlined in the ARRIVE Checklist to ensure transparency and rigor in reporting. All protocols, including sample collection procedures, were executed with explicit authorization from the animal owners and under the supervision of institutional ethics standards. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Out of Africa: genetic characterization and diversity of Mashona cattle in the United States.J Anim Sci. 2025 Jan 4;103:skaf045. doi: 10.1093/jas/skaf045. J Anim Sci. 2025. PMID: 39951431
-
Genomic Diversity and Selection Signatures for Zaosheng Cattle.Biology (Basel). 2025 May 28;14(6):623. doi: 10.3390/biology14060623. Biology (Basel). 2025. PMID: 40563874 Free PMC article.
-
Whole genome resequencing reveals genetic diversity, population structure, and selection signatures in local duck breeds.BMC Genomics. 2025 Aug 8;26(1):734. doi: 10.1186/s12864-025-11782-9. BMC Genomics. 2025. PMID: 40781581 Free PMC article.
-
Candidate Genes, Markers, Signatures of Selection, and Quantitative Trait Loci (QTLs) and Their Association with Economic Traits in Livestock: Genomic Insights and Selection.Int J Mol Sci. 2025 Aug 8;26(16):7688. doi: 10.3390/ijms26167688. Int J Mol Sci. 2025. PMID: 40869008 Free PMC article. Review.
-
Advances in the genetic characterization of guinea fowl in Africa: a comprehensive overview of the current status, progress of genome genotyping, and future perspectives.Poult Sci. 2025 May 28;104(9):105363. doi: 10.1016/j.psj.2025.105363. Online ahead of print. Poult Sci. 2025. PMID: 40482533 Free PMC article. Review.
References
-
- Mei C, Junjvlieke Z, RAZA S H A, et al. Copy number variation detection in Chinese Indigenous cattle by whole genome sequencing [J]. Genomics. 2020;112(1):831–6. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

