Comprehensive genomic study of Plasmodium falciparum in Sierra leone: genetic variation and resistance patterns
- PMID: 40597624
- PMCID: PMC12210811
- DOI: 10.1186/s12864-025-11771-y
Comprehensive genomic study of Plasmodium falciparum in Sierra leone: genetic variation and resistance patterns
Abstract
Background: Malaria remains a significant global health concern, with Plasmodium falciparum being the most dangerous of the malaria-causing parasites. Sierra Leone, with year-round transmission of malaria, continues to experience high infection rates, largely due to P. falciparum. Although genomic databases have provided valuable insights into malaria transmission patterns, drug resistance, and population structure, data from Sierra Leone has been limited. This study aims to build on our previous report by incorporating new samples and providing a more comprehensive genomic analysis of P. falciparum in Sierra Leone, with a particular focus on genetic diversity, population structure, and drug resistance.
Methods:
We collected P. falciparum samples from Freetown, Sierra Leone, between December 2022 and March 2023. A total of 35 samples underwent sequencing using the MGISEQ and Illumina platforms, resulting in high-coverage genomic data. Population structure was assessed using PCA, NJ trees, and STRUCTURE analysis, alongside comparisons with a global dataset from the pf3k project. Genetic diversity was evaluated using metrics such as ,
ω, Tajima’s D, and iHS. XPEHH was used to examine selection pressures across different regions.
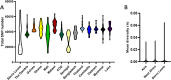
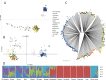
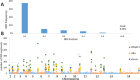
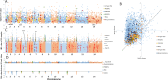
Results: Sequencing yielded over 376,450 high-quality SNPs across 35 Sierra Leone isolates, indicating substantial genetic variability. PCA, NJ trees, and STRUCTURE analysis revealed that the Sierra Leone isolates formed distinct clusters from both West African and Southeast Asian samples, with evidence of region-specific genetic adaptation. The low IBD we found suggested that infections were largely independent. Meanwhile the Tajima’s D and iHS analysis showed strong directional selection in genes associated with immune evasion and drug resistance genes, exhibiting ongoing evolutionary pressure due to therapeutic behavior.
Conclusion: This study provides us not only a genomic database but also a profile of P. falciparum in Sierra Leone, revealing high genetic diversity, strong selection pressure on drug resistance genes, and unique population structures. Our data emphasize the need for continued genomic surveillance to better manage malaria.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12864-025-11771-y.
Keywords: Drug resistance; Genomic diversity; Plasmodium falciparum; Population structure; Sierra Leone.
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Genome Analysis of Plasmodium falciparum: A Preliminary Observation - Sierra Leone, 2022-2023.China CDC Wkly. 2024 Apr 26;6(17):368-373. doi: 10.46234/ccdcw2024.071. China CDC Wkly. 2024. PMID: 38737823 Free PMC article.
-
Trends, and patterns, of premarital sexual intercourse and its associated factors among never-married young women aged 15-24 in Sierra Leone.PLoS One. 2024 Aug 22;19(8):e0309200. doi: 10.1371/journal.pone.0309200. eCollection 2024. PLoS One. 2024. PMID: 39172925 Free PMC article.
-
Assessment of copy number variation in genes related to drug resistance in Plasmodium vivax and Plasmodium falciparum isolates from the Brazilian Amazon and a systematic review of the literature.Malar J. 2017 Apr 19;16(1):152. doi: 10.1186/s12936-017-1806-z. Malar J. 2017. PMID: 28420389 Free PMC article.
-
Sex-disaggregated analysis of breast cancer prevalence in Sierra Leone.BMC Cancer. 2025 Jul 1;25(1):1022. doi: 10.1186/s12885-025-14403-6. BMC Cancer. 2025. PMID: 40597999 Free PMC article.
-
The link between the West African Ebola outbreak and health systems in Guinea, Liberia and Sierra Leone: a systematic review.Global Health. 2017 Jan 4;13(1):1. doi: 10.1186/s12992-016-0224-2. Global Health. 2017. PMID: 28049495 Free PMC article.
References
-
- World Health Organization. World malaria report 2023. World Health Organization; 2023.
-
- WHO guidelines for malaria [internet]. Geneva: World Health Organization; 2021. PMID: 36580567. - PubMed
Grants and funding
- TDRC-2019-194-30/National Sharing Service Platform for Parasite Resources
- TDRC-2019-194-30/National Sharing Service Platform for Parasite Resources
- TDRC-2019-194-30/National Sharing Service Platform for Parasite Resources
- GWVI-11.1-12/Three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai (2023-2025) Key Discipline Project
- GWVI-11.1-12/Three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai (2023-2025) Key Discipline Project
- GWVI-11.1-12/Three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai (2023-2025) Key Discipline Project
- GWVI-11.1-12/Three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai (2023-2025) Key Discipline Project
- 2020YFE0205700/National Research and Development Plan of China
- 2020YFE0205700/National Research and Development Plan of China
- 2020YFE0205700/National Research and Development Plan of China
- 2020YFE0205700/National Research and Development Plan of China
- INV-003421/GATES/Gates Foundation/United States
- INV-003421/GATES/Gates Foundation/United States
LinkOut - more resources
Full Text Sources

