Evidence of early genomic selection in Holstein Friesian across African and European ecosystems
- PMID: 40597625
- PMCID: PMC12211335
- DOI: 10.1186/s12864-025-11828-y
Evidence of early genomic selection in Holstein Friesian across African and European ecosystems
Abstract
Background: The Holstein Friesian (HF) cattle breed is the most dominant breed in commercial dairy farming worldwide and managed in more than 150 countries. These countries span diverse agro-climatic zones, ranging from tropical to cold regions. The introduction of HF animals in these regions occurred at different moments in the past which are poorly recorded and continued through importation of live animal and frozen semen. We hypothesize that the HF cattle populations in these regions underwent early forms of adaptation to these specific local environments. However, the detection of genetic variation associated with this adaptation remains poorly documented.
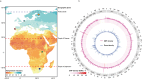
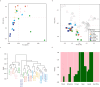
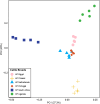
Results: This study investigates genetic relationship and potential early selection signatures in HF populations from three African countries (Egypt, South Africa, Uganda) and three European countries (Finland, Portugal, The Netherlands), considering five animals per country. Approximately 16.0 million single nucleotide polymorphisms (SNPs) were detected in the 30 HF animals and used for further analyses.
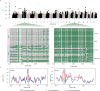
Across all countries, we identified dispersed regions totaling 3.3 megabase of ecosystem-specific genomic regions (43 genes), indicative of early selection signatures based on fixation indices (F-statistic, Fst). Furthermore, comparing variants between tropical (Egypt and Uganda) and cold regions (Finland and The Netherlands) by Fst, nucleotide diversity (θπ ratio), and extended haplotype homozygosity (XP-EHH), we identified a total of 10 candidate regions, comprising 12 genes within a 0.57 megabase size. The regions were enriched with genes involved in signaling pathways associated directly or indirectly with adaptation, including the immune system (PGLYRP4,PGLYRP3, PAG1, CD48, SLAMF1, DYSF,and LOC615223), organ development and reproduction (LDB3, ADAMTSL4, TPRN, CCDC40, OR2AG1G, and OR8B3), thermogenic activation (TBC1D16), phospholipid metabolism (PLPPR4 and PITPNB), thermos-tolerance (ZNF423), and stimulus response (NCOA7, CYP2C85, and ARFGEF3).
Conclusion: This study provides new insights into early forms of genetic plasticity of animals adapted to very diverse ecosystems. Our findings highlight candidate genes related to immune response, organ development, reproduction, metabolism, and thermo-tolerance, hypothesizing their role in facilitating adaptation to different environments.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12864-025-11828-y.
Keywords: Adaptation; Dairy farming; Holstein Friesian; Selective sweeps; Whole-genome resequencing.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All methods were performed in accordance with relevant guidelines and regulations. Blood samples were collected during the animals’ annual health inspections, conducted by licensed veterinarians. Prior to sample collection, written informed consent was obtained from each animal’s owner. The Egyptian Holstein Friesian blood sampling was done based on animal welfare guidelines of Institutional Animal Care and Use Committee, Cairo University (CU-IACUC) which approved this protocol under number CUIIF720. In Finland, animal handling procedures and sample collections were performed in accordance with the legislation approved by Regional State Administrative Agency for Southern Finland (ESAVI/31854/2019). In South Africa, sampling of blood and hair was performed with the approval of the Animal Ethics Committee of the Agricultural Research Council (APAEC [2020/17]), according to guidelines for the proper handling of animals during sample collection. In Uganda, the study was approved under Approval No: SBLS/HDRC/20/001 by the Research and Ethics Committee of the College of Veterinary Medicine, Animal Resources and Biosecurity (COVAB). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
The genomic comparison between autochthonous and cosmopolitan cows reveals structural variants involved in environmental adaptation.Sci Rep. 2025 Jul 1;15(1):22280. doi: 10.1038/s41598-025-07165-5. Sci Rep. 2025. PMID: 40593057 Free PMC article.
-
Breeding for organic dairy farming: what types of cows are needed?J Dairy Res. 2019 Feb;86(1):3-12. doi: 10.1017/S0022029919000141. J Dairy Res. 2019. PMID: 30907720 Review.
-
Differences in the expression of genes involved in the somatotropic axis in divergent strains of Holstein-Friesian dairy cows during early and mid lactation.J Dairy Sci. 2009 Oct;92(10):5229-38. doi: 10.3168/jds.2008-1856. J Dairy Sci. 2009. PMID: 19762841
-
Carcass characteristics of cattle differing in Jersey proportion.J Dairy Sci. 2018 Dec;101(12):11052-11060. doi: 10.3168/jds.2018-14992. Epub 2018 Sep 27. J Dairy Sci. 2018. PMID: 30268620
-
Biodiversity Genomics Research Practices Require Harmonising to Meet Stakeholder Needs in Conservation.Mol Ecol. 2025 Jun 26:e70001. doi: 10.1111/mec.70001. Online ahead of print. Mol Ecol. 2025. PMID: 40574266 Review.
References
-
- Houghton FL. Holstein-Friesian cattle: A history of the breed and its development in America: Press of the Holstein-Friesian Register; 1897.
-
- Ormiston E. Breeds of dairy cattle. Circular (University of Illinois (Urbana-Champaign campus) Extension Service in Agriculture and Home Economics); 543 1942.
-
- Hiemstra SJ, de Haas Y, Mäki-Tanila A, Gandini G. Local cattle breeds in europe: development of policies and strategies for self-sustaining breeds. Wageningen Academic; 2010.
Grants and funding
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- LEAP-Agri-326/Long-term EU-Africa Research and Innovation Partnership on Food and Nutrition Security and Sustainable Agriculture
- 202208610017/China Scholarship Council
- 2018/WOTRO/00488849/the Netherlands Organization for Scientistic Research
- 2018/WOTRO/00488849/the Netherlands Organization for Scientistic Research
- 2018/WOTRO/00488849/the Netherlands Organization for Scientistic Research
- 2018/WOTRO/00488849/the Netherlands Organization for Scientistic Research
- 2018/WOTRO/00488849/the Netherlands Organization for Scientistic Research
- 2018/WOTRO/00488849/the Netherlands Organization for Scientistic Research
- 727715/European Union's Horizon 2020 Research and Innovation Program
- 727715/European Union's Horizon 2020 Research and Innovation Program
- 727715/European Union's Horizon 2020 Research and Innovation Program
- 727715/European Union's Horizon 2020 Research and Innovation Program
- 727715/European Union's Horizon 2020 Research and Innovation Program
- 727715/European Union's Horizon 2020 Research and Innovation Program
- 319987/Research Council of Finland
- 2020.02754.CEECIND/CP1601/CP1649/CT0008/Fundação Nacional para a Ciência e a Tecnologia
- LEAP-Agri 326/Science, Technology & Innovation Funding Authority
- MoSTI/LEAP-11/Ministry of Science, Technology and Innovations
- 115577/National Research Foundation
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

