Integrated proteomics and transcriptomics analysis reveals key regulatory genes between ER-positive/PR-positive and ER-positive/PR-negative breast cancer
- PMID: 40597935
- PMCID: PMC12211939
- DOI: 10.1186/s12885-025-14451-y
Integrated proteomics and transcriptomics analysis reveals key regulatory genes between ER-positive/PR-positive and ER-positive/PR-negative breast cancer
Abstract
Purpose: Hormone receptor-positive breast cancer is characterized by the expression of estrogen receptor (ER) or progesterone receptor (PR), it is generally associated with less aggressive clinical features and more favorable prognostic outcomes, primarily due to the effectiveness of endocrine therapy. However, the loss of PR expression has been correlated with endocrine resistance and poorer prognosis. To date, there is limited research elucidating the underlying mechanisms distinguishing ER-positive/PR-positive from ER-positive/PR-negative breast cancer. This study aims to investigate the molecular mechanisms associated with these two subtypes and to propose recommendations for precision therapy.
Patients and methods: Fresh tumor tissues from ER + /PR + patients (n = 5) and ER + /PR- patients (n = 5) were subjected to proteomic analysis to identify differentially expressed proteins. Transcriptomic data were obtained from the TCGA database, encompassing 937 breast cancer patients divided into three subgroups: ER + /PR + (n = 627), ER + /PR- (n = 112), and ER-/PR- (n = 198). Clinical characteristics and prognostic data were collected to analyze disease-specific survival (DSS) and overall survival (OS) across the three subtypes. Differential expression data for both transcripts and proteins were extracted, and Cox regression along with Least Absolute Shrinkage and Selection Operator (LASSO) regression were applied to identify key regulatory genes. A risk scoring formula was employed to classify patients into high-risk and low-risk groups. Kaplan-Meier curves, Gene Set Enrichment Analysis (GSEA), immune cell infiltration analysis, and OncoPredict drug sensitivity predictions were conducted to provide insights into the underlying mechanisms and clinical treatment strategies for this patient cohort. The accuracy of this model was further validated using external GEO datasets (GSE21653, GSE20685, and GSE42568). Additionally, we collected data from 97 hormone receptor-positive breast cancer patients who underwent neoadjuvant chemotherapy at our center between January 2021 and December 2023, assessing their response to chemotherapy using the Miller-Payne score.
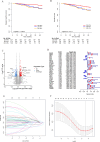
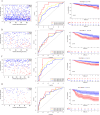
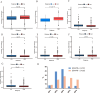
Results: In the TCGA database, patients with ER + /PR- breast cancer exhibited poorer 5-year DSS and OS compared to those with ER + /PR + status (DSS: P = 0.038; OS: P = 0.052), which was similar to those with ER-/PR- status (DSS: P = 0.47; OS: P = 0.77). 186 differentially expressed proteins (110 up-regulated and 76 down-regulated) were identified based on proteomic analysis. After COX regression and Lasso regression, five key differential genes with prognostic and diagnostic value of ER + /PR + and ER + /PR- patients were finally included, that is HPN, FSCN1, FGD3, LRIG1, and TBC1D7. HPN, FSCN1 and FGD3 can be regarded as a tumor suppressor gene. And LRIG1, and TBC1D7 can be regarded as a risk-associated gene. Patients with high-risk scores had significantly lower survival probabilities compared to those with low-risk scores. Additionally, there were differences in functional pathway enrichment analysis (galactose_metabolism, glycolysis_gluconeogenesis, jak_stat_signaling_pathway, pentose_phosphate_pathway, et al.) and immune cell infiltration (CD8 T cell, Macrophages M1, et al.) between the high-risk and low-risk groups. Drug sensitivity analysis indicated that the low-risk patients may be more sensitive to endocrine drug like fulvestrant, while high-risk patients may be more sensitive to chemotherapy drugs like docetaxel, paclitaxel, and vinorelbine. Of the 97 patients underwent neoadjuvant chemotherapy in our center, the proportion of patients achieving Miller-Payne (MP) score 4 and 5 was higher in ER + /PR- patients (44%) compared to ER + /PR + patients (9.8%).
Conclusion: We confirmed that ER + /PR- breast cancer patients exhibited worse survival compared with ER + /PR + patients. Five key regulatory genes were identified and potential mechanisms and biological pathways were discovered, our prediction of drug sensitivity offers new insights for developing precise pharmacological treatment strategies for ER + /PR- breast cancer.
Keywords: Estrogen receptor; Progesterone receptor; Prognosis; Proteomic analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval consent to participate: All experiments were performed in accordance with relevant guidelines and regulations. All experiments were performed in accordance with relevant guidelines and regulations. The studies involving human participants were reviewed and approved by Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University. The patients/participants provided their written informed consent to participate in this study. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
The Association between ER, PR, HER2, and ER-/PR+ Expression and Lung Cancer Subsequent in Breast Cancer Patients: A Retrospective Cohort Study Based on SEER Database.Breast J. 2023 Nov 11;2023:7028189. doi: 10.1155/2023/7028189. eCollection 2023. Breast J. 2023. PMID: 38021219 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Bioinformatics identification and validation of m6A/m1A/m5C/m7G/ac4 C-modified genes in oral squamous cell carcinoma.BMC Cancer. 2025 Jul 1;25(1):1055. doi: 10.1186/s12885-025-14216-7. BMC Cancer. 2025. PMID: 40597017 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Jin X, Zhou YF, Jiang YZ, Shao ZM. Molecular classification of hormone receptor-positive HER2-negative breast cancer. Nat Genet. 2023;55(10):1696–708. - PubMed
-
- Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. - PubMed
-
- Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: from early detection to new therapies. Radiologia. 2017;59(5):368–79. - PubMed
-
- Taraborrelli S. Physiology, production and action of progesterone. Acta Obstet Gynecol Scand. 2015;94(Suppl 161):8–16. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

